GENOMIC
Mapping
5q14.1. View the map and BAC contig (data from UCSC genome browser).
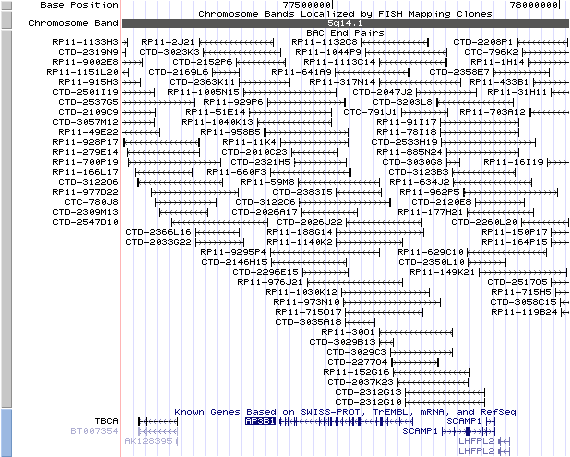
Structure
(assembly 07/03)
AP3B1/NM_003664: 27 exons, 292,378 bp, chr5:77,382,224-77,674,601.
The figure below shows the structure of the AP3B1 gene (data from UCSC genome browser).
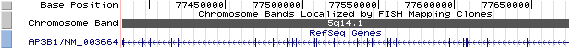
Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
AP3B1/NM_003664: 4,009bp, view ORF and the alignment to genomic.
Note: This RefSeq entry is based on AP3B1 entry BC038444 (4021bp), which matches a transcript (gi: 24638436), but exhibits differences between BC038444 and the human genome build 34. The encoded two proteins present one amino acid difference (E585V).
Expression Pattern
Tissue specificity: ubiquitous. Highest and lowest expression in kidney and liver tissue respectively.
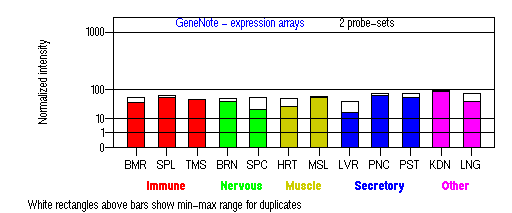
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle; LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
Beta3A-adaptin (NP_003655): 1094aa, ExPaSy NiceProt view of Swiss-Prot:O00203.
Synonyms: Adapter-related protein complex 3 beta 1 subunit; Adaptor protein complex AP-3 beta-1 subunit; AP-3 complex beta-1 subunit; Clathrin assembly protein complex 3 beta-1 large chain.
Ortholog
| Species | Mouse | Dog | Rat | Fruitfly | Yeast |
| GeneView | pe/Ap3b1 | AY221640 | Ap3b1 | CG11427 (rb) | APL6 |
| Protein | NP_033810 (1105aa) | AAP45786 (1091aa) | XM_226666 (1171aa) | AAF71924 (1160aa) | Apl6p (809aa) |
| Identities | 86% /965aa | 93% /1020aa | 82% /972aa | 49% /569 | 27% /223 |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) low complexity: 277 - 296
b) low complexity: 677 - 733
c) low complexity: 756 - 806
d) low complexity: 852 - 862
(2) Transmembrane domains predicted by SOSUI: none.
(3) Pfam domains: PF01602 - Adaptin N terminal region.
(4) Graphic view of InterPro domain structure.
(5) CDD domain: KOG1060: Vesicle coat complex AP-3, beta subunit [Intracellular trafficking, secretion, and vesicular transport].
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site [pattern] [Warning: pattern with a high probability of occurrence]:
75 - 78 NASE, 401 - 404 NIST, 907 - 910 NNTT, 908 - 911 NTTD, 1004 - 1007 NETS.
b) Tyrosine sulfation site [rule] [Warning: rule with a high probability of occurrence]:
267 - 281 edngknfYesdddqk.
c) cAMP- and cGMP-dependent protein kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
741 - 744 KRnS, 795 - 798 RRvT. Evidence shown the protein is phosphorylated on serine residues.
d) Tyrosine kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
344 - 351 RsnrEvq.Y
e) GLU_RICH Glutamic acid-rich region [profile]:
678 - 802.
f) SER_RICH Serine-rich region [profile]:
677-794.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: found KLPI at 923
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: too long tail
i) Dileucine motif in the tail: found, LL at 114.
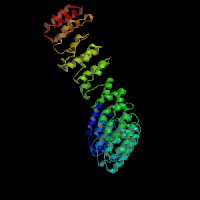
3D Model
ModBase: predicted comparative 3D structures on O00203 (data from UCSC Gene Sorter). (from left to right: Front, Top, Side view)
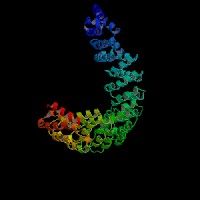
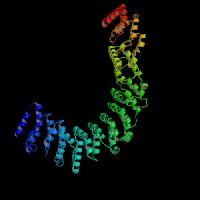
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=121,320Da, pI=5.75 (NP_003655).
FUNCTION
Ontology
a) Biological process: intracellular protein transport (overview of trafficking pathway here).
b) Biological process: endocytsis
c) Component of Golgi apparatus.
d) Plays a role in protein sorting in the late-Golgi/trans-Golgi network (TGN) and/or endosomes.
e) Plays a role in the vesicular trafficking of tyrosinase to melanosomes (view diagram of melanosomal protein sorting here).
Location
Component of the coat surrounding the cytoplasmic face of coated vesicles located at the Golgi complex.
Interaction
The HPS2/AP3B1 gene encodes the adapter-related protein complex 3 beta 1 subunit (beta-adaptin 3A, or AP-3 complex beta-3A subunit). The AP-3 complex is a heterotetramer composed of two large adaptins (delta/AP3D1 and beta3A/AP3B2 or beta3B/AP3B1), a medium adaptin (mu3A/AP3M1 or mu3B/AP3M2) and a small adaptin (sigma3A/AP3S1 or sigma3B/AP3S2). The AP3M1 subunit interacts with tyrosinase for lysosomal targeting (Honing, et al). Sugita, et al showed that CD1B, but not other CD1 isoforms, binds the AP3 adaptor protein complex. AP-3 binds to a PAR1 C-tail localized tyrosine-based motif and mediates PAR1 lysosomal degradation. Moreover, AP-3 facilitates PAR1 interaction with ALIX (Dores, et al). More cargoes for AP-3 complex are reviewed by Dell'Angelica (2009).
6 proteins are shown to be associated with APL6 in Yeast GRID.
AP3B1 drosophila homolog CG11427 interaction information in CuraGen interaction database.
Pathway
Lysosomes. AP-3 complex is associated with the Golgi region as well as more peripheral structures. It facilitates the budding of vesicles from the Golgi membrane and appears to be involved in the sorting of a subset of transmembrane proteins targeted to lysosomes and lysosome-related organelles (view diagram of AP-3 pathway here).Vps class C/HOPS subunits and clathrin exist in complex with either AP-3 or hepatocyte growth factor receptor substrate (Hrs) (Zlatic, et al). AP-3 delta subunit binds Vps41 of HOPS complex directly (Angers and Merz AJ). HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway (Wilkin, et al). BLOC-1, PI4KIIalpha, and AP-3 belong to a tripartite complex in lysosomal trafficking (Salazar, et al).
Melanosomes. It is proposed that AP-3 facilitates membrane recruitment of BLOC-1, which in turn facilitates AP-3 (and BLOC-2) dissociation. It seems that BLOC-1 might serve as a tethering factor downstream of AP-3-mediated vesicle formation (Di Pietro, et al). Both AP-1- and AP-3-favoring OCA2 variants required BLOC-1 for melanosomal transport. BLOC-1 can cooperate with either adaptor during cargo sorting to LROs (Sitaram, et al).
Synaptic vesicles. AP-3 and BLOC-1 function in sorting both synaptic vesicle membrane proteins (such as ZnT3) and characteristic AP-3 lysosomal cargoes (such as LAMP1, PI4KIIalpha). Lysosomal and lysosome-related organelle biogenesis mechanisms regulate steady-state synaptic vesicle protein composition from shared early endosomes (Newell-Litwa, et al (2009)). AP-3 and BLOC-1 differentially regulate the composition of presynaptic terminals in the striatum and dentate gyrus of the hippocampus (Newell-Litwa, et al (2010)). AP-3B is expressed in islets and mediates beta-cell synaptic-like microvesicles (SLMVs) biogenesis (Suckow, et al).
Immune cells. Sugita, et al proposed that there is an AP3-dependent pathway for antigen presentation by CD1B molecules.
MUTATION
Allele or SNP
Mutational alleles described in HGMD.
Allelic variants described in OMIM.
SNPs deposited in dbSNP.
Distribution
Location
Genomic
cDNA
Protein
Type
Strain
Reference
Exon 2
153_156del 4bp
153_156del 4bp
E52del 4bp
frame-shift
64X
Caucasian
Wenham, et al
Exon 8
904A>T
904A>T
R302X
nonsense
Dutch
Enders, et al
Exon 10
1062delCA ins TATCAATATC
1062delCA ins TATCAATATC
Q355delCA ins TATCAATATC
frame-shift, 360X
Italian
Fontana, et al
Exon 12
1166_1128del 63bp
1166_1128del 63bp
L390_Q410del 63bp
in-frame
Dutch
Dell'Angelica, et al
Intron 14
splicing donor, +6 T>C
1473ins 39bp
T491ins 39bp
frame-shift, 496X
English
Clark, et al
Exon 15
1525C>T
1525C>T
R509X
nonsense
Cajun/Houma Indian
Huizing, et al (2002)
Exon 15
1618insG
1618insG
G540insG
frame-shift, 565X
English
Clark, et al
Exon 16
1739T>G
1739T>G
L580R
missense
Dutch
Dell'Angelica, et al
Exon 16
1789insA
1789insA
I597insA
frame-shift,608X
Italian
Fontana, et al
Exon 18
1975G>T
1975G>T
E659X
nonsense
Cajun/Houma Indian
Huizing, et al (2002)
Intron 18+
Exon 19
180242_180866del
2078_2165del 88bp
E693del 88bp
frame-shift, 707X
Caucasian
Wenham, et al
Exon 27
3222_3223delTG
3222_3223delTG
T1074delTG
frame-shift, 1135X
Dutch
Kurnik, et al
(Numbering of genomic and cDNA sequence is based on the start codon of RefSeq NM_003664.)
A clinically diagnosed HPS2 patient showed a homozygous pericentric inv(5)(p15.1q14.1), which disrupts the AP3B1 gene (Jones, et al).
Effect
Most of the reported patients are compound heterozygotes except one patient with homozygous R302X mutation. The nonsense mutations (R509X and E659X) produce no mRNA and no AP3B1 protein (Huizing, et al (2002) ). The double heterozygous mutations (Δ 390-410 and L580R) in two brothers produce normal amounts of AP3B1 mRNA but exhibite drastically reduced levels of AP3 due to enhanced degradation of mutant beta-3A (Dell'Angelica, et al ). The patient with the splice site mutation (T>C) and G540insG might have a very small amount of wild-type protein, but they could not find beta3A, gamma, or mu3A, three subunits of the AP-3 complex , indicating that loss of the beta3A subunit induces instability of the complex and rapid degradation of other subunits (Clark, et al). The two patients with a compound mutation (Q355fsX4; I597fsX10) show a complete absence of beta-3A (Fontana, et al). The mutation (c.3222_3223delTG) in the last exon of AP3B1 causes a frameshift and a prolonged altered protein (T1074fsX60). The location of the deletion at the very C-terminal end may prevent a complete loss of the HPS2 protein leading to a less pronounced severity of immunodeficiency than in other HPS2 patients (Kurnik, et al).
PHENOTYPE
Defects in AP3B1 are the cause of Hermansky-Pudlak syndrome type 2 (HPS-2, OMIM 608233 ) (Dell'Angelica, et al). All HPS-2 patients so far reported have oculocutaneous albinism, absent platelet dense bodies, and distinguishing feature of neutropenia. HPS-2 differs from the other forms of HPS in that it includes immunodeficiency in its phenotype and patients with HPS-2 have an increased susceptibility to infections. The degranulation defect of CTL and NK cells impairs lysis of targeted cells. Mild facial dysmorphia, hepatosplenomegaly, developmental delay, and pulmonary fibrosis have been reported (Huizing, et al (2002) ). HPS-2 patients had findings of interstitial lung disease (ILD) on a high-resolution computed tomography scan of the chest,with elevated TGFbeta1 and IL-17A correlated with the severity (Gochuico, et al). Hemophagocytic lymphohistocytosis (HLH), which represents lysosomal trafficking disorder, has also been reported in HPS-2 patients (Enders, et al; Ma, et al). However, HPS2 confers a risk for HLH that is lower than in Griscelli or Chediak-Higashi syndrome, probably because of a milder defect in cytotoxicity (Jessen, et al).
AP-3 complex has been widely shown to be involved in the biogenesis of lysosome-related organelles, such as melanosomes and platelet dense granules. One particular role of AP-3 is its involvement in the granule biogenesis of immune cells. This leads to the hallmark of AP-3 in innate immunity, antigen presentation and CTL killing. In addition, AP-3 mediates many cargoes for lysosomal targeting, such as LAMPs, VAMP7-TI, PI4KIIalpha. Failure of lysosomal degradation often leads to the increase of membrane proteins on cell surface. For example, HPS-2 fibrablasts exhibit increased surface expression (mislocalization) of lysosomal proteins (e.g. LAMP-1, LAMP-2, and LAMP-3) through the plasma membrane (Dell'Angelica, et al). HPS-2 melanocytes showed tyrosinase was restricted to the perinuclear region (Huizing, et al (2001) ). Misorting of lysosomal proteins such as CD63 or CD107 to the cell membrane has been found on fibroblasts, neutrophils or CTLs. Neutrophil elastase (NE) and perforin content are reduced in HPS-2 patients, and the cytolytic activity of NK cells are impaired. These suggest the impaired innate immunity in HPS-2 (Fontana, et al; Jung, et al). NE appears to be one of the cargo proteins of AP-3 complex (Horwitz, et al).
In AP3-deficient cells from patients with HPS2, CD1B failed to gain access to lysosomes efficiently and was mislocalized to the plasma membrane and early endosomes. The failure in CD1B trafficking resulted in a profound failure to present microbial lipid antigens efficiently. The defects in CD1B antigen presentation may account for the recurrent bacterial infections in HPS2 patients (Sugita, et al) (view diagram of CD1B blockage in APC cells here). In dendritic cells (DCs), AP-3 is involved in efficient TLR recruitment to phagosomes and MHC-II presentation of antigens internalized by phagocytosis but not receptor-mediated endocytosis. In AP-3 deficient DCs, peptide:MHC-II export to the cell surface was impeded (Mantegazza, et al). AP-3, as well as the BLOC-1 and BLOC-2 are essential for plasmacytoid dendritic cells (pDCs) signaling through TLR7 and TLR9. However, these proteins are not necessary for TLR7 or TLR9 signaling in conventional DCs (Blasius, et al). By studying CD8-positive cytotoxic T lymphocytes (CTLs) from an HPS2 patient, Clark, et al determined that AP-3 deficiency results in loss of microtubule-mediated movement of enlarged perforin- and granzyme-containing lytic granules toward the immunologic synapse and a profound loss of CTL-mediated killing (view diagram of lytic granule blockage in CTL cells here). It has shown that AP-3 complex is required for HIV-1 assembly and release (Liu, et al).
AP-3 functions in neurons to mediate protein trafficking to synaptic vesicles and modulate cell surface receptor density. AP3B1 is a unbiquitously expressed isoform, while AP3B2 is a neuronal-specific isoform (Newell-Litwa, et al (2007)). The functions of AP3B1 and AP3B2 containing complexes are distinct and divergent (Seong, et al).. A direct physical interaction between M5 and the AP-3 adaptor complex regulator AGAP1 was identified. This interaction mediated the binding of AP-3 to M5 which is required for the cell surface receptor density after sustained receptor stimulation (Bendor, et al).
REFERENCE
- Angers CG, Merz AJ. HOPS interacts with Apl5 at the vacuole membrane and is required for consumption of AP-3 transport vesicles. Mol Biol Cell 2009; 20: 4563-74. PMID: 19741093
- Bendor J, Lizardi-Ortiz JE, Westphalen RI, Brandstetter M, Hemmings HC Jr, Sulzer D, Flajolet M, Greengard P. AGAP1/AP-3-dependent endocytic recycling of M5 muscarinic receptors promotes dopamine release. EMBO J 2010; 29: 2813-26. PMID: 20664521
- Blasius AL, Arnold CN, Georgel P, Rutschmann S, Xia Y, Lin P, Ross C, Li X, Smart NG, Beutler B. Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci U S A 2010; 107: 19973-8. PMID: 21045126
- Clark RH, Stinchcombe JC, Day A, Blott E, Booth S, Bossi G, Hamblin T, Davies EG, Griffiths GM. Adaptor protein 3-dependent microtubule-mediated movement of lytic granules to the immunological synapse. Nat Immunol 2003; 4: 1111-20.
PMID : 14566336
- Dell'Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Cell Biol 2009; 21: 552-9. Review. PMID: 19497727
- Dell'Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell 1999; 3: 11-21. PMID: 10024875
- Di Pietro SM, Falcon-Perez JM, Tenza D, et al. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell 2006; 17:4027-38. PMID: 16837549
- Dores MR, Paing MM, Lin H, Montagne WA, Marchese A, Trejo J. AP-3 regulates PAR1 ubiquitin-independent MVB/lysosomal sorting via an ALIX-mediated pathway. Mol Biol Cell 2012; 23: 3612-23. PMID: 22833563
- Enders A, Zieger B, Schwarz K, Yoshimi A, Speckmann C, Knoepfle EM, Kontny U, Muller C, Nurden A, Rohr J, Henschen M, Pannicke U, Niemeyer C, Nurden P, Ehl S. Lethal hemophagocytic lymphohistiocytosis in Hermansky-Pudlak Syndrome Type II. Blood 2006; 108: 81-7. PMID: 16551969
- Fontana S, Parolini S, Vermi W, Booth S, Gallo F, Donini M, Benassi M, Gentili F, Ferrari D, Notarangelo LD, Cavadini P, Marcenaro E, Dusi S, Cassatella M, Facchetti F, Griffiths GM, Moretta A, Notarangelo LD, Badolato R. Innate immunity defects in Hermansky-Pudlak type 2 syndrome. Blood 2006; 107: 4857-64.PMID: 16507770
- Gochuico BR, Huizing M, Golas GA, Scher CD, Tsokos M, Denver SD, Frei-Jones MJ, Gahl WA. Interstitial lung disease and pulmonary fibrosis in Hermansky-Pudlak syndrome type 2, an adaptor protein-3 complex disease. Mol Med 2012; 18: 56-64. PMID: 22009278
- Honing S, Sandoval IV, von Figura K. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J 1998; 17: 1304-14. PMID: 9482728
- Horwitz MS, Duan Z, Korkmaz B, Lee HH, Mealiffe ME, Salipante SJ. Neutrophil elastase in cyclic and severe congenital neutropenia.Blood 2007; 109: 1817-24. PMID: 17053055
- Huizing M, Sarangarajan R, Strovel E, Zhao Y, Gahl WA, Boissy RE. AP-3 mediates tyrosinase but not TRP-1 trafficking in human melanocytes. Mol Biol Cell 2001; 12: 2075-85. PMID: 11452004
- Huizing M, Scher CD, Strovel E, Fitzpatrick DL, Hartnell LM, Anikster Y, Gahl WA. Nonsense mutations in ADTB3A cause complete deficiency of the beta3A subunit of adaptor complex-3 and severe Hermansky-Pudlak syndrome type 2. Pediatr Res 2002; 51: 150-8. PMID: 11809908
- Jessen B, Bode SF, Ammann S, Chakravorty S, Davies G, Diestelhorst J, Frei-Jones M, Gahl WA, Gochuico BR, Griese M, Griffiths G, Janka G, Klein C, Kogl T, Kurnik K, Lehmberg K, Maul-Pavicic A, Mumford AD, Pace D, Parvaneh N, Rezaei N, de Saint Basile G, Schmitt-Graeff A, Schwarz K, Karasu GT, Zieger B, Zur Stadt U, Aichele P, Ehl S. The risk of hemophagocytic lymphohistiocytosis in Hermansky-Pudlak syndrome type 2. Blood 2013; 121: 2943-51. PMID: 23403622
- Jones ML, Murden SL, Brooks C, Maloney V, Manning RA, Gilmour KC, Bharadwaj V, de la Fuente J, Chakravorty S, Mumford AD. Disruption of AP3B1 by a chromosome 5 inversion: a new disease mechanism in Hermansky-Pudlak syndrome type 2. BMC Med Genet 2013; 14: 42. PMID: 23557002
- Jung J, Bohn G, Allroth A, Boztug K, Brandes G, Sandrock I, Schaffer AA, Rathinam C, Kollner I, Beger C, Schilke R, Welte K, Grimbacher B, Klein C. Identification of a homozygous deletion in the AP3B1 gene causing Hermansky-Pudlak syndrome, type 2. Blood 2006; 108: 362-9.PMID: 16537806
- Kurnik K, Bartsch I, Maul-Pavicic A, Ehl S, Sandrock-Lang K, Bidlingmaier C, Rombach N, Busse A, Belohradsky BH, Muller-Hocker J, Aslanidis C, Schmitz G, Zieger B. Novel mutation in Hermansky-Pudlak syndrome type 2 with mild immunological phenotype. Platelets 2012; [Epub ahead of print] PMID: 23215637
- Liu L, Sutton J, Woodruff E, Villalta F, Spearman P, Dong X. Defective HIV-1 particle assembly in AP-3-deficient cells derived from patients with Hermansky-Pudlak syndrome type 2. J Virol 2012; 86: 11242-53. PMID: 22875976
- Ma D, Rudd E, Edner J, Gavhed S, Ramme KG, Fadeel B, Nordenskjold M, Henter JI, Zheng C. Sequence analysis of the SRGN, AP3B1, ARF6, and SH2D1A genes in familial hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer 2008; 50: 1067-9.
PMID: 18000860
- Mantegazza AR, Guttentag SH, El-Benna J, Sasai M, Iwasaki A, Shen H, Laufer TM, Marks MS. Adaptor protein-3 in dendritic cells facilitates phagosomal toll-like receptor signaling and antigen presentation to CD4(+) T cells. Immunity 2012; 36: 782-94. PMID: 22560444
- Newell-Litwa K, Chintala S, Jenkins S, Pare JF, McGaha L, Smith Y, Faundez V. Hermansky-Pudlak protein complexes, AP-3 and BLOC-1, differentially regulate presynaptic composition in the striatum and hippocampus. J Neurosci 2010; 30: 820-31. PMID: 20089890
- Newell-Litwa K, Salazar G, Smith Y, Faundez V. Roles of BLOC-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles. Mol Biol Cell 2009; 20: 1441-53. PMID: 19144828
- Newell-Litwa K, Seong E, Burmeister M, Faundez V. Neuronal and non-neuronal functions of the AP-3 sorting machinery. J Cell Sci 2007; 120: 531-41. Review. PMID: 17287392
- Salazar G, Zlatic S, Craige B, Peden AA, Pohl J, Faundez V. Hermansky-Pudlak syndrome protein complexes associate with phosphatidylinositol 4-kinase type II alpha in neuronal and non-neuronal cells. J Biol Chem 2009; 284: 1790-802. PMID: 19010779
- Seong E, Wainer BH, Hughes ED, Saunders TL, Burmeister M, Faundez V.
Genetic analysis of the neuronal and ubiquitous AP-3 adaptor complexes reveals divergent functions in brain. Mol Biol Cell 2005; 16: 128-40. PMID: 15537701
- Sitaram A, Dennis MK, Chaudhuri R, De Jesus-Rojas W, Tenza D, Setty SR, Wood CS, Sviderskaya EV, Bennett DC, Raposo G, Bonifacino JS, Marks MS. Differential recognition of a dileucine-based sorting signal by AP-1 and AP-3 reveals a requirement for both BLOC-1 and AP-3 in delivery of OCA2 to melanosomes. Mol Biol Cell 2012; 23: 3178-92. PMID: 22718909
- Suckow AT, Craige B, Faundez V, Cain WJ, Chessler SD. An AP-3-dependent mechanism drives synaptic-like microvesicle biogenesis in pancreatic islet beta-cells. Am J Physiol Endocrinol Metab 2010; 299: E23-32. PMID: 20442321
- Sugita M, Cao X, Watts GF, Rogers RA, Bonifacino JS, Brenner MB. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity 2002; 16: 697-706. PMID: 12049721
- Wenham M, Grieve S, Cummins M, Jones ML, Booth S, Kilner R, Ancliff PJ, Griffiths GM, Mumford AD. Two patients with Hermansky Pudlak syndrome type 2 and novel mutations in AP3B1. Haematologica 2010; 95: 333-7. PMID: 19679886
- Zlatic SA, Tornieri K, L'Hernault SW, Faundez V. Clathrin-dependent mechanisms modulate the subcellular distribution of class C Vps/HOPS tether subunits in polarized and nonpolarized cells. Mol Biol Cell 2011; 22: 1699-715. PMID: 21411634
EDIT HISTORY:
Created by Wei Li: 06/21/2004
Updated by Wei Li: 04/05/2006
Updated by Wei Li: 12/25/2006
Updated by Wei Li: 10/08/2007
Updated by Wei Li: 02/29/2008
Updated by Wei Li: 05/25/2011
Updated by Wei Li: 07/29/2012
Updated by Wei Li: 06/13/2013
(1) Predicted results by ScanProsite:
a) N-glycosylation site [pattern] [Warning: pattern with a high probability of occurrence]:
75 - 78 NASE, 401 - 404 NIST, 907 - 910 NNTT, 908 - 911 NTTD, 1004 - 1007 NETS.
b) Tyrosine sulfation site [rule] [Warning: rule with a high probability of occurrence]:
267 - 281 edngknfYesdddqk.
c) cAMP- and cGMP-dependent protein kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
741 - 744 KRnS, 795 - 798 RRvT. Evidence shown the protein is phosphorylated on serine residues.
d) Tyrosine kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
344 - 351 RsnrEvq.Y
e) GLU_RICH Glutamic acid-rich region [profile]:
678 - 802.
f) SER_RICH Serine-rich region [profile]:
677-794.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: found KLPI at 923
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: too long tail
i) Dileucine motif in the tail: found, LL at 114.
3D Model
ModBase: predicted comparative 3D structures on O00203 (data from UCSC Gene Sorter). (from left to right: Front, Top, Side view)



2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=121,320Da, pI=5.75 (NP_003655).
ModBase: predicted comparative 3D structures on O00203 (data from UCSC Gene Sorter). (from left to right: Front, Top, Side view)



2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=121,320Da, pI=5.75 (NP_003655).
FUNCTION
Ontology
a) Biological process: intracellular protein transport (overview of trafficking pathway here).
b) Biological process: endocytsis
c) Component of Golgi apparatus.
d) Plays a role in protein sorting in the late-Golgi/trans-Golgi network (TGN) and/or endosomes.
e) Plays a role in the vesicular trafficking of tyrosinase to melanosomes (view diagram of melanosomal protein sorting here).
Location
Component of the coat surrounding the cytoplasmic face of coated vesicles located at the Golgi complex.
Interaction
The HPS2/AP3B1 gene encodes the adapter-related protein complex 3 beta 1 subunit (beta-adaptin 3A, or AP-3 complex beta-3A subunit). The AP-3 complex is a heterotetramer composed of two large adaptins (delta/AP3D1 and beta3A/AP3B2 or beta3B/AP3B1), a medium adaptin (mu3A/AP3M1 or mu3B/AP3M2) and a small adaptin (sigma3A/AP3S1 or sigma3B/AP3S2). The AP3M1 subunit interacts with tyrosinase for lysosomal targeting (Honing, et al). Sugita, et al showed that CD1B, but not other CD1 isoforms, binds the AP3 adaptor protein complex. AP-3 binds to a PAR1 C-tail localized tyrosine-based motif and mediates PAR1 lysosomal degradation. Moreover, AP-3 facilitates PAR1 interaction with ALIX (Dores, et al). More cargoes for AP-3 complex are reviewed by Dell'Angelica (2009).
6 proteins are shown to be associated with APL6 in Yeast GRID.
AP3B1 drosophila homolog CG11427 interaction information in CuraGen interaction database.
Pathway
Lysosomes. AP-3 complex is associated with the Golgi region as well as more peripheral structures. It facilitates the budding of vesicles from the Golgi membrane and appears to be involved in the sorting of a subset of transmembrane proteins targeted to lysosomes and lysosome-related organelles (view diagram of AP-3 pathway here).Vps class C/HOPS subunits and clathrin exist in complex with either AP-3 or hepatocyte growth factor receptor substrate (Hrs) (Zlatic, et al). AP-3 delta subunit binds Vps41 of HOPS complex directly (Angers and Merz AJ). HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway (Wilkin, et al). BLOC-1, PI4KIIalpha, and AP-3 belong to a tripartite complex in lysosomal trafficking (Salazar, et al).
Melanosomes. It is proposed that AP-3 facilitates membrane recruitment of BLOC-1, which in turn facilitates AP-3 (and BLOC-2) dissociation. It seems that BLOC-1 might serve as a tethering factor downstream of AP-3-mediated vesicle formation (Di Pietro, et al). Both AP-1- and AP-3-favoring OCA2 variants required BLOC-1 for melanosomal transport. BLOC-1 can cooperate with either adaptor during cargo sorting to LROs (Sitaram, et al).
Synaptic vesicles. AP-3 and BLOC-1 function in sorting both synaptic vesicle membrane proteins (such as ZnT3) and characteristic AP-3 lysosomal cargoes (such as LAMP1, PI4KIIalpha). Lysosomal and lysosome-related organelle biogenesis mechanisms regulate steady-state synaptic vesicle protein composition from shared early endosomes (Newell-Litwa, et al (2009)). AP-3 and BLOC-1 differentially regulate the composition of presynaptic terminals in the striatum and dentate gyrus of the hippocampus (Newell-Litwa, et al (2010)). AP-3B is expressed in islets and mediates beta-cell synaptic-like microvesicles (SLMVs) biogenesis (Suckow, et al).
Immune cells. Sugita, et al proposed that there is an AP3-dependent pathway for antigen presentation by CD1B molecules.
MUTATION
Allele or SNP
Mutational alleles described in HGMD.
Allelic variants described in OMIM.
SNPs deposited in dbSNP.
Distribution
| Location | Genomic | cDNA | Protein | Type | Strain | Reference |
| Exon 2 | 153_156del 4bp | 153_156del 4bp | E52del 4bp | frame-shift 64X |
Caucasian | Wenham, et al |
| Exon 8 | 904A>T | 904A>T | R302X | nonsense | Dutch | Enders, et al |
| Exon 10 | 1062delCA ins TATCAATATC | 1062delCA ins TATCAATATC | Q355delCA ins TATCAATATC | frame-shift, 360X | Italian | Fontana, et al |
| Exon 12 | 1166_1128del 63bp | 1166_1128del 63bp | L390_Q410del 63bp | in-frame | Dutch | Dell'Angelica, et al |
| Intron 14 | splicing donor, +6 T>C | 1473ins 39bp | T491ins 39bp | frame-shift, 496X | English | Clark, et al |
| Exon 15 | 1525C>T | 1525C>T | R509X | nonsense | Cajun/Houma Indian | Huizing, et al (2002) |
| Exon 15 | 1618insG | 1618insG | G540insG | frame-shift, 565X | English | Clark, et al |
| Exon 16 | 1739T>G | 1739T>G | L580R | missense | Dutch | Dell'Angelica, et al |
| Exon 16 | 1789insA | 1789insA | I597insA | frame-shift,608X | Italian | Fontana, et al |
| Exon 18 | 1975G>T | 1975G>T | E659X | nonsense | Cajun/Houma Indian | Huizing, et al (2002) |
| Intron 18+ Exon 19 |
180242_180866del | 2078_2165del 88bp | E693del 88bp | frame-shift, 707X | Caucasian | Wenham, et al |
| Exon 27 | 3222_3223delTG | 3222_3223delTG | T1074delTG | frame-shift, 1135X | Dutch | Kurnik, et al |
A clinically diagnosed HPS2 patient showed a homozygous pericentric inv(5)(p15.1q14.1), which disrupts the AP3B1 gene (Jones, et al).
Effect
Most of the reported patients are compound heterozygotes except one patient with homozygous R302X mutation. The nonsense mutations (R509X and E659X) produce no mRNA and no AP3B1 protein (Huizing, et al (2002) ). The double heterozygous mutations (Δ 390-410 and L580R) in two brothers produce normal amounts of AP3B1 mRNA but exhibite drastically reduced levels of AP3 due to enhanced degradation of mutant beta-3A (Dell'Angelica, et al ). The patient with the splice site mutation (T>C) and G540insG might have a very small amount of wild-type protein, but they could not find beta3A, gamma, or mu3A, three subunits of the AP-3 complex , indicating that loss of the beta3A subunit induces instability of the complex and rapid degradation of other subunits (Clark, et al). The two patients with a compound mutation (Q355fsX4; I597fsX10) show a complete absence of beta-3A (Fontana, et al). The mutation (c.3222_3223delTG) in the last exon of AP3B1 causes a frameshift and a prolonged altered protein (T1074fsX60). The location of the deletion at the very C-terminal end may prevent a complete loss of the HPS2 protein leading to a less pronounced severity of immunodeficiency than in other HPS2 patients (Kurnik, et al).
PHENOTYPE
Defects in AP3B1 are the cause of Hermansky-Pudlak syndrome type 2 (HPS-2, OMIM 608233 ) (Dell'Angelica, et al). All HPS-2 patients so far reported have oculocutaneous albinism, absent platelet dense bodies, and distinguishing feature of neutropenia. HPS-2 differs from the other forms of HPS in that it includes immunodeficiency in its phenotype and patients with HPS-2 have an increased susceptibility to infections. The degranulation defect of CTL and NK cells impairs lysis of targeted cells. Mild facial dysmorphia, hepatosplenomegaly, developmental delay, and pulmonary fibrosis have been reported (Huizing, et al (2002) ). HPS-2 patients had findings of interstitial lung disease (ILD) on a high-resolution computed tomography scan of the chest,with elevated TGFbeta1 and IL-17A correlated with the severity (Gochuico, et al). Hemophagocytic lymphohistocytosis (HLH), which represents lysosomal trafficking disorder, has also been reported in HPS-2 patients (Enders, et al; Ma, et al). However, HPS2 confers a risk for HLH that is lower than in Griscelli or Chediak-Higashi syndrome, probably because of a milder defect in cytotoxicity (Jessen, et al).
AP-3 complex has been widely shown to be involved in the biogenesis of lysosome-related organelles, such as melanosomes and platelet dense granules. One particular role of AP-3 is its involvement in the granule biogenesis of immune cells. This leads to the hallmark of AP-3 in innate immunity, antigen presentation and CTL killing. In addition, AP-3 mediates many cargoes for lysosomal targeting, such as LAMPs, VAMP7-TI, PI4KIIalpha. Failure of lysosomal degradation often leads to the increase of membrane proteins on cell surface. For example, HPS-2 fibrablasts exhibit increased surface expression (mislocalization) of lysosomal proteins (e.g. LAMP-1, LAMP-2, and LAMP-3) through the plasma membrane (Dell'Angelica, et al). HPS-2 melanocytes showed tyrosinase was restricted to the perinuclear region (Huizing, et al (2001) ). Misorting of lysosomal proteins such as CD63 or CD107 to the cell membrane has been found on fibroblasts, neutrophils or CTLs. Neutrophil elastase (NE) and perforin content are reduced in HPS-2 patients, and the cytolytic activity of NK cells are impaired. These suggest the impaired innate immunity in HPS-2 (Fontana, et al; Jung, et al). NE appears to be one of the cargo proteins of AP-3 complex (Horwitz, et al).
In AP3-deficient cells from patients with HPS2, CD1B failed to gain access to lysosomes efficiently and was mislocalized to the plasma membrane and early endosomes. The failure in CD1B trafficking resulted in a profound failure to present microbial lipid antigens efficiently. The defects in CD1B antigen presentation may account for the recurrent bacterial infections in HPS2 patients (Sugita, et al) (view diagram of CD1B blockage in APC cells here). In dendritic cells (DCs), AP-3 is involved in efficient TLR recruitment to phagosomes and MHC-II presentation of antigens internalized by phagocytosis but not receptor-mediated endocytosis. In AP-3 deficient DCs, peptide:MHC-II export to the cell surface was impeded (Mantegazza, et al). AP-3, as well as the BLOC-1 and BLOC-2 are essential for plasmacytoid dendritic cells (pDCs) signaling through TLR7 and TLR9. However, these proteins are not necessary for TLR7 or TLR9 signaling in conventional DCs (Blasius, et al). By studying CD8-positive cytotoxic T lymphocytes (CTLs) from an HPS2 patient, Clark, et al determined that AP-3 deficiency results in loss of microtubule-mediated movement of enlarged perforin- and granzyme-containing lytic granules toward the immunologic synapse and a profound loss of CTL-mediated killing (view diagram of lytic granule blockage in CTL cells here). It has shown that AP-3 complex is required for HIV-1 assembly and release (Liu, et al).
AP-3 functions in neurons to mediate protein trafficking to synaptic vesicles and modulate cell surface receptor density. AP3B1 is a unbiquitously expressed isoform, while AP3B2 is a neuronal-specific isoform (Newell-Litwa, et al (2007)). The functions of AP3B1 and AP3B2 containing complexes are distinct and divergent (Seong, et al).. A direct physical interaction between M5 and the AP-3 adaptor complex regulator AGAP1 was identified. This interaction mediated the binding of AP-3 to M5 which is required for the cell surface receptor density after sustained receptor stimulation (Bendor, et al).
REFERENCE
- Angers CG, Merz AJ. HOPS interacts with Apl5 at the vacuole membrane and is required for consumption of AP-3 transport vesicles. Mol Biol Cell 2009; 20: 4563-74. PMID: 19741093
- Bendor J, Lizardi-Ortiz JE, Westphalen RI, Brandstetter M, Hemmings HC Jr, Sulzer D, Flajolet M, Greengard P. AGAP1/AP-3-dependent endocytic recycling of M5 muscarinic receptors promotes dopamine release. EMBO J 2010; 29: 2813-26. PMID: 20664521
- Blasius AL, Arnold CN, Georgel P, Rutschmann S, Xia Y, Lin P, Ross C, Li X, Smart NG, Beutler B. Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci U S A 2010; 107: 19973-8. PMID: 21045126
- Clark RH, Stinchcombe JC, Day A, Blott E, Booth S, Bossi G, Hamblin T, Davies EG, Griffiths GM. Adaptor protein 3-dependent microtubule-mediated movement of lytic granules to the immunological synapse. Nat Immunol 2003; 4: 1111-20. PMID : 14566336
- Dell'Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Cell Biol 2009; 21: 552-9. Review. PMID: 19497727
- Dell'Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell 1999; 3: 11-21. PMID: 10024875
- Di Pietro SM, Falcon-Perez JM, Tenza D, et al. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell 2006; 17:4027-38. PMID: 16837549
- Dores MR, Paing MM, Lin H, Montagne WA, Marchese A, Trejo J. AP-3 regulates PAR1 ubiquitin-independent MVB/lysosomal sorting via an ALIX-mediated pathway. Mol Biol Cell 2012; 23: 3612-23. PMID: 22833563
- Enders A, Zieger B, Schwarz K, Yoshimi A, Speckmann C, Knoepfle EM, Kontny U, Muller C, Nurden A, Rohr J, Henschen M, Pannicke U, Niemeyer C, Nurden P, Ehl S. Lethal hemophagocytic lymphohistiocytosis in Hermansky-Pudlak Syndrome Type II. Blood 2006; 108: 81-7. PMID: 16551969
- Fontana S, Parolini S, Vermi W, Booth S, Gallo F, Donini M, Benassi M, Gentili F, Ferrari D, Notarangelo LD, Cavadini P, Marcenaro E, Dusi S, Cassatella M, Facchetti F, Griffiths GM, Moretta A, Notarangelo LD, Badolato R. Innate immunity defects in Hermansky-Pudlak type 2 syndrome. Blood 2006; 107: 4857-64.PMID: 16507770
- Gochuico BR, Huizing M, Golas GA, Scher CD, Tsokos M, Denver SD, Frei-Jones MJ, Gahl WA. Interstitial lung disease and pulmonary fibrosis in Hermansky-Pudlak syndrome type 2, an adaptor protein-3 complex disease. Mol Med 2012; 18: 56-64. PMID: 22009278
- Honing S, Sandoval IV, von Figura K. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J 1998; 17: 1304-14. PMID: 9482728
- Horwitz MS, Duan Z, Korkmaz B, Lee HH, Mealiffe ME, Salipante SJ. Neutrophil elastase in cyclic and severe congenital neutropenia.Blood 2007; 109: 1817-24. PMID: 17053055
- Huizing M, Sarangarajan R, Strovel E, Zhao Y, Gahl WA, Boissy RE. AP-3 mediates tyrosinase but not TRP-1 trafficking in human melanocytes. Mol Biol Cell 2001; 12: 2075-85. PMID: 11452004
- Huizing M, Scher CD, Strovel E, Fitzpatrick DL, Hartnell LM, Anikster Y, Gahl WA. Nonsense mutations in ADTB3A cause complete deficiency of the beta3A subunit of adaptor complex-3 and severe Hermansky-Pudlak syndrome type 2. Pediatr Res 2002; 51: 150-8. PMID: 11809908
- Jessen B, Bode SF, Ammann S, Chakravorty S, Davies G, Diestelhorst J, Frei-Jones M, Gahl WA, Gochuico BR, Griese M, Griffiths G, Janka G, Klein C, Kogl T, Kurnik K, Lehmberg K, Maul-Pavicic A, Mumford AD, Pace D, Parvaneh N, Rezaei N, de Saint Basile G, Schmitt-Graeff A, Schwarz K, Karasu GT, Zieger B, Zur Stadt U, Aichele P, Ehl S. The risk of hemophagocytic lymphohistiocytosis in Hermansky-Pudlak syndrome type 2. Blood 2013; 121: 2943-51. PMID: 23403622
- Jones ML, Murden SL, Brooks C, Maloney V, Manning RA, Gilmour KC, Bharadwaj V, de la Fuente J, Chakravorty S, Mumford AD. Disruption of AP3B1 by a chromosome 5 inversion: a new disease mechanism in Hermansky-Pudlak syndrome type 2. BMC Med Genet 2013; 14: 42. PMID: 23557002
- Jung J, Bohn G, Allroth A, Boztug K, Brandes G, Sandrock I, Schaffer AA, Rathinam C, Kollner I, Beger C, Schilke R, Welte K, Grimbacher B, Klein C. Identification of a homozygous deletion in the AP3B1 gene causing Hermansky-Pudlak syndrome, type 2. Blood 2006; 108: 362-9.PMID: 16537806
- Kurnik K, Bartsch I, Maul-Pavicic A, Ehl S, Sandrock-Lang K, Bidlingmaier C, Rombach N, Busse A, Belohradsky BH, Muller-Hocker J, Aslanidis C, Schmitz G, Zieger B. Novel mutation in Hermansky-Pudlak syndrome type 2 with mild immunological phenotype. Platelets 2012; [Epub ahead of print] PMID: 23215637
- Liu L, Sutton J, Woodruff E, Villalta F, Spearman P, Dong X. Defective HIV-1 particle assembly in AP-3-deficient cells derived from patients with Hermansky-Pudlak syndrome type 2. J Virol 2012; 86: 11242-53. PMID: 22875976
- Ma D, Rudd E, Edner J, Gavhed S, Ramme KG, Fadeel B, Nordenskjold M, Henter JI, Zheng C. Sequence analysis of the SRGN, AP3B1, ARF6, and SH2D1A genes in familial hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer 2008; 50: 1067-9. PMID: 18000860
- Mantegazza AR, Guttentag SH, El-Benna J, Sasai M, Iwasaki A, Shen H, Laufer TM, Marks MS. Adaptor protein-3 in dendritic cells facilitates phagosomal toll-like receptor signaling and antigen presentation to CD4(+) T cells. Immunity 2012; 36: 782-94. PMID: 22560444
- Newell-Litwa K, Chintala S, Jenkins S, Pare JF, McGaha L, Smith Y, Faundez V. Hermansky-Pudlak protein complexes, AP-3 and BLOC-1, differentially regulate presynaptic composition in the striatum and hippocampus. J Neurosci 2010; 30: 820-31. PMID: 20089890
- Newell-Litwa K, Salazar G, Smith Y, Faundez V. Roles of BLOC-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles. Mol Biol Cell 2009; 20: 1441-53. PMID: 19144828
- Newell-Litwa K, Seong E, Burmeister M, Faundez V. Neuronal and non-neuronal functions of the AP-3 sorting machinery. J Cell Sci 2007; 120: 531-41. Review. PMID: 17287392
- Salazar G, Zlatic S, Craige B, Peden AA, Pohl J, Faundez V. Hermansky-Pudlak syndrome protein complexes associate with phosphatidylinositol 4-kinase type II alpha in neuronal and non-neuronal cells. J Biol Chem 2009; 284: 1790-802. PMID: 19010779
- Seong E, Wainer BH, Hughes ED, Saunders TL, Burmeister M, Faundez V. Genetic analysis of the neuronal and ubiquitous AP-3 adaptor complexes reveals divergent functions in brain. Mol Biol Cell 2005; 16: 128-40. PMID: 15537701
- Sitaram A, Dennis MK, Chaudhuri R, De Jesus-Rojas W, Tenza D, Setty SR, Wood CS, Sviderskaya EV, Bennett DC, Raposo G, Bonifacino JS, Marks MS. Differential recognition of a dileucine-based sorting signal by AP-1 and AP-3 reveals a requirement for both BLOC-1 and AP-3 in delivery of OCA2 to melanosomes. Mol Biol Cell 2012; 23: 3178-92. PMID: 22718909
- Suckow AT, Craige B, Faundez V, Cain WJ, Chessler SD. An AP-3-dependent mechanism drives synaptic-like microvesicle biogenesis in pancreatic islet beta-cells. Am J Physiol Endocrinol Metab 2010; 299: E23-32. PMID: 20442321
- Sugita M, Cao X, Watts GF, Rogers RA, Bonifacino JS, Brenner MB. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity 2002; 16: 697-706. PMID: 12049721
- Wenham M, Grieve S, Cummins M, Jones ML, Booth S, Kilner R, Ancliff PJ, Griffiths GM, Mumford AD. Two patients with Hermansky Pudlak syndrome type 2 and novel mutations in AP3B1. Haematologica 2010; 95: 333-7. PMID: 19679886
- Zlatic SA, Tornieri K, L'Hernault SW, Faundez V. Clathrin-dependent mechanisms modulate the subcellular distribution of class C Vps/HOPS tether subunits in polarized and nonpolarized cells. Mol Biol Cell 2011; 22: 1699-715. PMID: 21411634
EDIT HISTORY:
Created by Wei Li: 06/21/2004
Updated by Wei Li: 04/05/2006
Updated by Wei Li: 12/25/2006
Updated by Wei Li: 10/08/2007
Updated by Wei Li: 02/29/2008
Updated by Wei Li: 05/25/2011
Updated by Wei Li: 07/29/2012
Updated by Wei Li: 06/13/2013