GENOMIC
Mapping
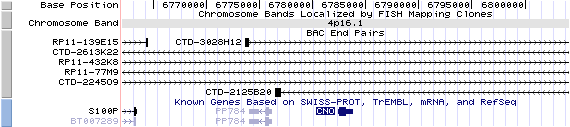
4p16.1. View the map and BAC contig (data from UCSC genome browser).
Structure
(assembly 7/03)
CNO/NM_018366: 1 exon (intronless), 1,546bp, Chr4: 6,782,383 - 6,783,786.
The figure below shows the structure of the CNO gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
CNO (NM_018366), 1,546bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: ubiquitously expressed.
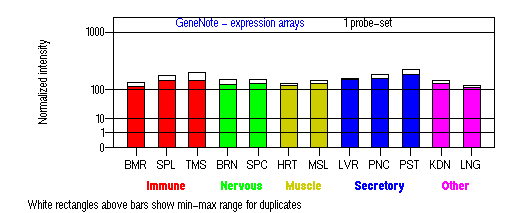
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle; LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
Cappuccino protein homolog (NP_060836): 217aa, ExPaSy NiceProt view of Swiss-Prot:Q9NUP1.
Ortholog
| Species | Mouse | Rat | Fugu fish | Mosquito | Fruit fly |
| GeneView | Cno | LOC364183 | 132443 | Q7PP24 | CG14149 |
| Protein | NP_598485 (215aa) | XP_344256 (215aa) | 140268 (210aa) | XP_315539 (184aa) | NP_648414 (169aa) |
| Identities | 77%/168aa | 77%/169aa | 39%/76aa | 31%/42aa | 27%/40aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
low complexity: 8 - 25
(2) Transmembrane domains predicted by SOSUI: none.
Motif/Site
(1) Predicted results by ScanProsite:
a) Casein kinase II phosphorylation site : [occurs frequently]
4 - 7: SfsD, 88 - 91: SleD, 94 - 97: TrvD, 133 - 136: SriD 157 - 160: TkaE.
b) N-myristoylation site : [occurs frequently]
23 - 28: GAawSG, 48 - 53: GAedGA, 146 - 151: GGrvAR.
c) Cell attachment sequence : [occurs frequently]
107 - 109: RGD.
,br>
d) Protein kinase C phosphorylation site : [occurs frequently]
212 - 214: SeR.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase: none.
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
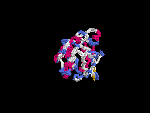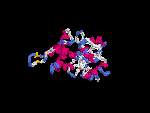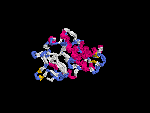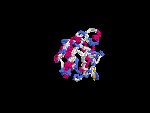
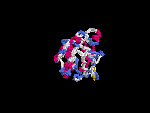
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=23,351Da, pI=4.90 (NP_060836).
FUNCTION
Ontology
Involved in the development of lysosome-related organelles, such as melanosomes and platelet-dense granules.
Location
Cytoplasmic.
Interaction
Interacts with pallidin, muted, dysbindin, and snapin directly(Ciciotte, et al; Li, et al (2004); Starcevic, et al). The Cno protein is a subunit of the biogenesis of lysosome-related organelles complex 1 (BLOC-1), in which it interacts with the products of seven other HPS genes, mu, pa, sdy, rp, Snapap, Blos1, Blos2 (Ciciotte, et al; Falcon-Perez , et al; Li, et al (2003); Moriyama, et al; Starcevic, et al) (view diagram of BLOC-1 complex here).
Cno drosophila homolog CG14149 interaction information in CuraGen interaction database.
Pathway
The encoded protein may play a role in intracellular vesicular trafficking. Nguyen, et al found that the greatest percentages of immature melanosomes were observed in the BLOC-1 mouse mutants pa and cno, which suggests the maturation of melanosome is blocked between the multivesicular body stage and stage I (view diagram of melanosome blockage here). The cappuccino gene encodes a product involved in an AP-3-independent mechanism critical to the biogenesis of lysosome-related organelles (Gwynn, et al). (view diagram of BLOC-1 and AP-3 pathway here)
MUTATION
Allele or SNP
Ciciotte, et al performed mutation screening of the human CNO gene in 18 HPS patients with no defect in previously identified HPS genes by amplification of genomic DNA from cultured fibroblasts and sequencing. No defects were observed.
SNPs deposited in dbSNP.
Distribution
(none)
Effect
(none)
PHENOTYPE
(Animal Models)
Mutation in the Cno gene is the cause of cappuccino (cno) mutant (Ciciotte, et al), a mouse model of Hermansky-Pudlak syndrome (OMIM 605695). Note that The mouse cappuccino gene is not related to the cappuccino (capu) gene of Drosophila.
REFERENCE
- Ciciotte SL, Gwynn B, Moriyama K, Huizing M, Gahl WA, Bonifacino JS, Peters LL. Cappuccino, a mouse model of Hermansky-Pudlak syndrome, encodes a novel protein that is part of the pallidin-muted complex (BLOC-1). Blood 2003; 101: 4402-7. PMID: 12576321
- Falcon-Perez JM, Starcevic M, Gautam R, Dell'Angelica EC. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem 2002; 277: 28191-9. PMID: 12019270
- Gwynn B, Ciciotte SL, Hunter SJ, Washburn LL, Smith RS, Andersen SG, Swank RT, Dell'Angelica EC, Bonifacino JS, Eicher EM, Peters LL. Defects in the cappuccino (cno) gene on mouse chromosome 5 and human 4p cause Hermansky-Pudlak syndrome by an AP-3-independent mechanism. Blood 2000; 96: 4227-35. PMID: 11110696
- Li W, Rusiniak ME, Chintala S, Gautam R, Novak EK, Swank RT. Murine Hermansky-Pudlak syndrome genes: regulators of lysosome-related organelles. Bioessays 2004; 26: 616-28. PMID: 15170859
- Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O'Brien EP, Tinsley CL, Blake DJ, Spritz RA, Copeland NG, Jenkins NA, Amato D, Roe BA, Starcevic M, Dell'Angelica EC, Elliott RW, Mishra V, Kingsmore SF, Paylor RE, Swank RT. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat Genet 2003; 35: 84-9. PMID: 12923531
- Moriyama K, Bonifacino JS. Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic 2002; 3: 666-77. PMID: 12191018
- Nguyen T, Novak EK, Kermani M, Fluhr J, Peters LL, Swank RT, Wei ML. Melanosome morphologies in murine models of hermansky-pudlak syndrome reflect blocks in organelle development. J Invest Dermatol 2002; 119: 1156-64. PMID: 12445206
- Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J Biol Chem 2004; 279: 28393-401. PMID: 15102850
EDIT HISTORY:
Created by Wei Li & Jonathan Bourne 07/01/2004