GENOMIC
Mapping
12q13.2. View the map and FISH BAC clones (data from UCSC genome browser).

Structure
(assembly 07/03)
BLOC1S1/NM_001487: 4 exons, 3,665 bp, chr12:54,396,087-54,399,751.
The figure below shows the structure of the BLOC1S1 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=human BLOC1S1, seq2=mouse Bloc1s1) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
BLOC1S1/NM_001487: 567 bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: Ubiquitously expressed (Inoue, et al). Northern-blot analysis revealed expression of 0.7-kb and 1.1-kb transcripts in all human tissues examined (Watanabe, et al).
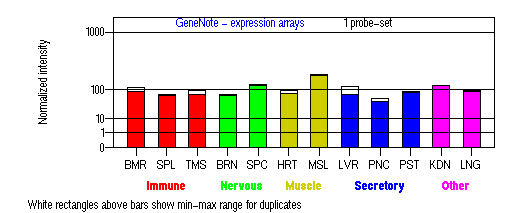
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle; LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
GCN5 like protein 1
(NP_001478): 125aa, ExPaSy NiceProt view of Swiss-Prot:P78537.
Synonym: RT14 protein.
Ortholog
| Species | Mouse | Rat | Zebrafish |
| GeneView | Bloc1s1 | RT14 | 14171 |
| Protein | NP_056555 (125aa) | XP_213833 (125aa) | 13120 (125aa) |
| Identities | 99%/125aa | 100%/125aa | 91%/123aa |
| Species | Drosophila | Mosquito | Worm |
| GeneView | CG30077 | 1281235 | 3L300 |
| Protein | NP_725401 (143aa) | XP_321175 (148aa) | NP_499262 (129aa) |
| Identities | 58%/113aa | 64%/117aa | 51%/120aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. Note that the human GCN5L1 protein displays weak homology (27%) to yeast GCN5, which is also called general control of amino-acid synthesis 5-like 1 (Driessen, et al ).
Domain
(1) Domains predicted by SMART:
a) low complexity: 12 - 26
b) coiled coil: 1-32
c) Pfam:GCN5L1: 5-125
(2) Transmembrane domains predicted by SOSUI: none.
(3) Pfam domain: Pfam:PF06320, GCN5L1.
(4) CDD domain: KOG3390.
(5) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-myristoylation site : [occurs frequently]
45 - 50: GVaqAY,
75 - 80: GQwiGM,
79 - 84: GMveNF,
118 - 123: GQlqSA.
b) Casein kinase II phosphorylation site : [occurs frequently]
110 - 113: TalE.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER membrane retention signals: XXRR-like motif in the N-terminus: LSRL
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase: none.
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
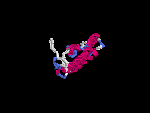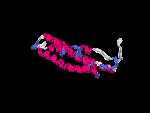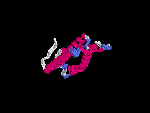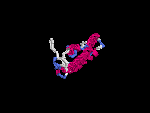
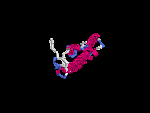
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=14,311Da, pI=7.88.
FUNCTION
Ontology
(1) May play a role in intracellular vesicle trafficking.
(2) Protein interaction in BLOC-1.
Location
Cytoplasmic.
Interaction
BLOC-1 subunit 1 is a subunit of the biogenesis of lysosome-related organelles complex 1 (BLOC-1), which contains the products of seven other HPS genes, DTNBP1, MU, PLDN, CNO, BLOS2, BLOS3, SNAPAP (Ciciotte, et al; Falcon-Perez , et al; Li, et al; Moriyama, et al; Starcevic, et al). It interacts with pallidin, snapin, and BLOS2 within the complex ( Starcevic, et al) (view diagram of BLOC-1 complex here). In Arabidopsis, BLOS1 interacts with SNX1 to modulate the vacuole degradation of auxin effluxers PIN1/2 to regulate root development ( Cui, et al).
BLOS1 drosophila homolog CG30077 interaction information in CuraGen interaction database.
Pathway
Involved in the development of lysosome-related organelles, such as melanosomes and platelet-dense granules (view diagram of BLOC-1 pathway here).
REFERENCE
- Ciciotte SL, Gwynn B, Moriyama K, Huizing M, Gahl WA, Bonifacino JS, Peters LL. Cappuccino, a mouse model of Hermansky-Pudlak syndrome, encodes a novel protein that is part of the pallidin-muted complex (BLOC-1). Blood 2003; 101: 4402-7. PMID: 12576321
- Cui Y, Li X, Chen Q, He X, Yang Q, Zhang A, Yu X, Chen H, Liu N, Xie Q, Yang W, Zuo J, Palme K, Li W. BLOS1, a putative BLOC-1 subunit, interacts with SNX1 and modulates root growth in Arabidopsis. J Cell Sci 2010; 123: 3727-33. PMID: 20971704
- Driessen CA, Winkens HJ, Kuhlmann LD, Janssen BP, van Vugt AH, Deutman AF, Janssen JJ. Cloning and structural analysis of the murine GCN5L1 gene. Gene 1997; 203: 27-31. PMID: 9426003
- Falcon-Perez JM, Starcevic M, Gautam R, Dell'Angelica EC. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem 2002; 277: 28191-9. PMID: 12019270
- Inoue M, Isomura M, Ikegawa S, Fujiwara T, Shin S, Moriya H, Nakamura Y. Isolation and characterization of a human cDNA clone (GCN5L1) homologous to GCN5, a yeast transcription activator. Cytogenet Cell Genet 1996; 73: 134-6. PMID: 8646881
- Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O'Brien EP, Tinsley CL, Blake DJ, Spritz RA, Copeland NG, Jenkins NA, Amato D, Roe BA, Starcevic M, Dell'Angelica EC, Elliott RW, Mishra V, Kingsmore SF, Paylor RE, Swank RT. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat Genet 2003; 35: 84-9. PMID: 12923531
- Moriyama K, Bonifacino JS. Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic 2002; 3: 666-77. PMID: 12191018
- Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J Biol Chem 2004; 279: 28393-401. PMID: 15102850
- Watanabe TK, Fujiwara T, Shinomiya H, Kuga Y, Hishigaki H, Nakamura Y, Hirai Y. Molecular cloning of a novel human cDNA, RT14, containing a putative ORF highly conserved between human, fruit fly, and nematode. DNA Res 1995; 2: 235-7. PMID: 8770567
EDIT HISTORY:
Created by Wei Li, 07/08/2004
Updated by Wei Li, 05/26/2011