GENOMIC
Mapping
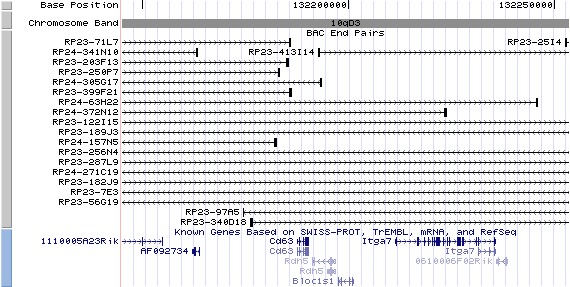
10qD3. View the map and BAC contig (data from UCSC genome browser).
>Structure
(assembly 10/03)
Bloc1s1/NM_015740: 4 exons, 3,612 bp, chr10:132,197,519-132,201,130.
The figure below shows the structure of the Bloc1s1 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=human BLOC1S1, seq2=mouse Bloc1s1) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Bloc1s1/NM_015740: 551 bp, view ORF and the alignment to genomic.
Expression Pattern
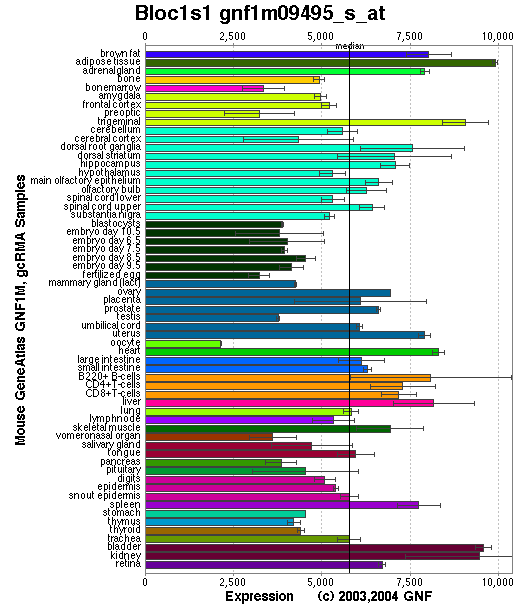
Tissue specificity: Ubiquitously expressed (by similarity). Highly expressed in the small intestine.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
General control of amino acid synthesis-like 1
(Gcn5l1/NP_056555): 125aa, ExPaSy NiceProt view of Swiss-Prot:Q9DC96.
Ortholog
| Species | Human | Rat | Zebrafish |
| GeneView | BLOC1S1 | RT14 | 14171 |
| Protein | NP_001478 (125aa) | XP_213833 (125aa) | 13120 (125aa) |
| Identities | 99%/125aa | 99%/125aa | 91%/123aa |
| Species | Drosophila | Mosquito | Worm |
| GeneView | CG30077 | 1281235 | 3L300 |
| Protein | NP_725401 (143aa) | XP_321175 (148aa) | NP_499262 (129aa) |
| Identities | 58%/113aa | 64%/117aa | 51%/120aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. Note that the murine Gcn5l1, like the human GCN5L1 protein, displays weak homology (27%) to yeast GCN5 (Driessen, et al ).
Domain
(1) Domains predicted by SMART:
a) low complexity: 12 - 26
b) coiled coil: 1-32
c) Pfam:GCN5L1: 5-125
(2) Transmembrane domains predicted by SOSUI: none.
(3) Pfam domain: Pfam:PF06320, GCN5L1.
(4) CDD domain: KOG3390.
(5) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-myristoylation site : [occurs frequently]
45 - 50: GVaqAY,
75 - 80: GQwiGM,
79 - 84: GMveNF,
118 - 123: GQlqSA.
b) Casein kinase II phosphorylation site : [occurs frequently]
110 - 113: TalE.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER membrane retention signals: XXRR-like motif in the N-terminus: LSRL
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase: none.
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.


2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=14,281Da, pI=7.88.
FUNCTION
Ontology
(1) May play a role in intracellular vesicle trafficking.
(2) Protein interaction in BLOC-1.
Location
Cytoplasmic.
Interaction
BLOC-1 subunit 1 is a subunit of the biogenesis of lysosome-related organelles complex 1 (BLOC-1), which contains the products of seven other HPS genes, sdy, mu, pa, cno, rp, Snapap, Blos2 (Ciciotte, et al; Falcon-Perez , et al; Li, et al; Moriyama, et al; Starcevic, et al). It interacts with pallidin, snapin, and Blos2 within the complex ( Starcevic, et al) (view diagram of BLOC-1 complex here).
Blos1 drosophila homolog CG30077 interaction information in CuraGen interaction database.
Pathway
Involved in the development of lysosome-related organelles, such as melanosomes and platelet-dense granules (view diagram of BLOC-1 pathway here).
MUTATION
Allele or SNP
No phenotypic allele is described in MGI:1195276.
No mutation has been reported in mouse mutants.
SNPs deposited in dbSNP.
Distribution
(none)
Effect
(none)
PHENOTYPE
(none)