GENOMIC
Mapping
14qC1 View the map and BAC contig (data from UCSC genome browser).
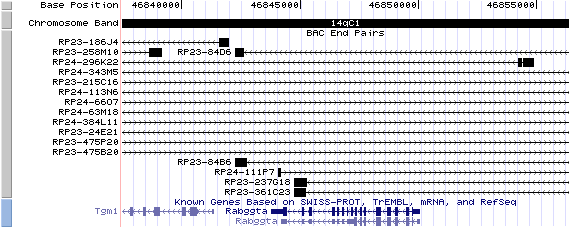
Structure
(assembly 10/03)
Rabggta/NM_019519: 16 exons, 2,547bp, Chr14: 46,843,772 - 46,850,071.
The figure below shows the structure of Rabggta (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Rabggta (NM_019519),
2,547bp, view ORF and the alignment to genomic..
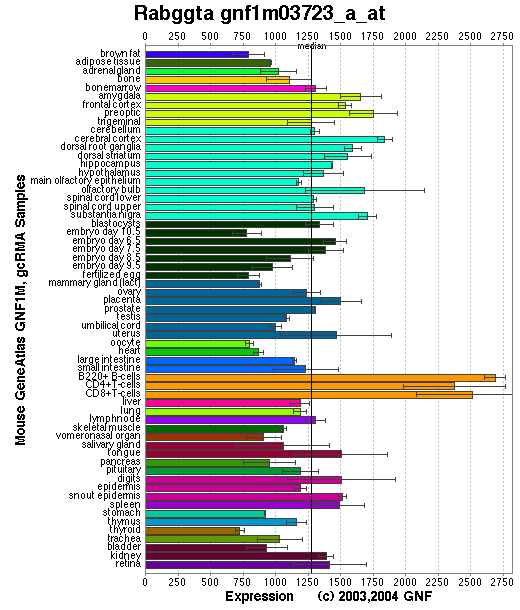
Expression Pattern
Tissue specificity: ubiquitous.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
Geranylgeranyl transferase type II alpha subunit (NP_062392): 567aa, ExPaSy NiceProt view of Swiss-Prot:Q9JHK4.
Synonyms: Rab geranylgeranyltransferase alpha subunit; Rab GG transferase alpha;
Rab GGTase alpha.
Ortholog
| Species | Human | Rat | Zebrafish | Worm |
| GeneView | RABGGTA | LOC58983 | 19427 | M57.2 |
| Protein | NP_878256 (567aa) | NP_113842 (567aa) | 7197 (333aa) |
CE19541 (580aa) |
| Identities | 90%/515aa | 95%/543aa | 53%/184aa | 31%/181aa |
| Species | Mosquito | Fruitfly | Yeast | |
| GeneView | 1279131 | CG12007 | YJL031C | |
| Protein | XP_318802 (495aa) | Q9VN77 (515aa) |
YJL031C (327aa) | |
| Identities | 39%/190aa | 38%/179aa | 31%/81aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) Pfam:PPTA repeats:
47 - 77; 91 - 121; 127 - 157; 162 - 192; 210-240.
b) Pfam:LRR: 462 - 485, 507 - 531.
(2) Graphical view of InterPro domain structure.
(3) Transmembrane domains predicted by SOSUI: none.
(4) CDD domains:
a) COG5536: Protein prenyltransferase, alpha subunit (post-translational modification).
b) COG0529: Protein geranylgeranyltransferase type II, alpha subunit (post-translational modification).
Motif/Site
(1) Predicted results by ScanProsite:
a) Leucine-rich region profile : [occurs frequently]
385 - 540: score=8.770
b) Protein kinase C phosphorylation site : [occurs frequently]
78 - 80: TqK, 317 - 319: TfR, 327 - 329: TqK, 370 - 372: ScK, 410 - 412: TlK
c) N-glycosylation site : [occurs frequently]
174 - 177: NFSN, 177 - 180: NYSS
d) N-myristoylation site : [occurs frequently]
268 - 273: GSkmGT, 542 - 547: GNslCQ
e) Tyrosine kinase phosphorylation site : [occurs frequently]
393 - 400: RalDpllY
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: XXRR-like motif in the N-terminus: HGRL
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase (Q9JHK4): predicted comparative 3D structures (data from UCSC Genome Sorter).
From left to right: Front, Top, and Side views of predicted protein.
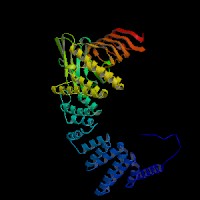
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
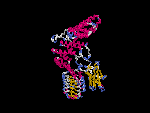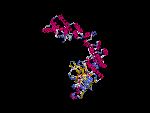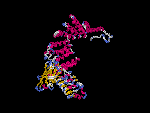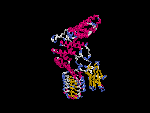
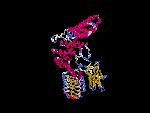
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=64,989Da, pI=5.54 (NP_062392).
FUNCTION
Ontology
a) Process: protein amino acid prenylation.
b) Function: protein prenyltransferase activity.
Location
Cytoplasmic.
Interaction
The Rabggta gene encode the geranylgeranyl transferase type II alpha subunit (EC 2.5.1.60) (Rab geranylgeranyltransferase alpha subunit, or RabGGTase alpha). Rab GGTase catalyzes the transfer of a geranyl-geranyl moiety from geranyl-geranyl pyrophosphate to both cysteines in Rab proteins with an -XXCC, -XCXC and -CCXX C-terminal, such as Rab1a, Rab3a, Rab5a, and Rab27a respectively. The enzymatic reaction requires the aid of a Rab escort protein (REP, also called component A). Heterodimer of an alpha and a beta subunit, is collectively called component B.
2 proteins are shown to be associated with YJL031C/BET4 in Yeast GRID.
Rabggta drosophila homolog CG12007 interaction information in CuraGen interaction database.
Pathway
The enzyme RabGGTase covalently attaches the geranylgeranyl groups to two C-terminal cysteines of the Rab protein. NiceZyme View of protein geranylgeranyltransferase type II within ENZYME: EC 2.5.1.60. More details in BRENDA: RabGGTase.
MUTATION
Allele or SNP
1 phenotypic allele is described in MGI: 1860443 .
SNPs deposited in dbSNP.
Distribution
| Location | Genomic | cDNA | Protein | Type | Strain | Reference |
| Exon 1 | splicing acceptor, -55G>A | (a) -54A~3Gdel 57bp, (b) -122G~-42Gdel 81bp, (c) -55G>A |
(a): no ATG, (b) and (c): normal |
splicing | gm (B6) | Detter, et al |
Effect
The net result of the mutation is a significant reduction in levels of RabGGTase alpha subunit protein and reduced levels of RabGGTase (Detter, et al ). Zhang, et al further observed that significant deficits in prenylation and membrane binding of most Rabs were observed in platelets and melanocytes (view the defects in melanocyte dynamics here). In contrast, minimal alterations in Rab prenylation were apparent in several other gunmetal tissues despite the fact that RabGGTase activity was equally diminished in these tissues (tissue-specific effects).
PHENOTYPE
Mutation in the Rabggta gene is the cause of gunmetal mutant (Detter, et al), a mouse model of Hermansky-Pudlak syndrome (OMIM 601905). The gm allele arose from C57BL/6J. Homozygotes for a spontaneous hypomorphic mutation exhibit diluted pigmentation, a platelet defect resulting in prolonged bleeding (due to reduced rates of platelet synthesis, abnormalities of platelet alpha and dense granules and hypopigmentation), macrothrombocytopenia, impaired killing by cytotoxic T lymphocytes, high mortality, and poor breeding (Novak, et al; Swank, et al). The mutant is described in more detail in JAX Mice database (C57BL/6J-Rabggtagm/J) and Mouse Locus Card #Rabggta.
Life span is severely reduced in the gunmetal mutants (McGarry, et al). Richards-Smith, et al found that the gunmetal (gm/gm) platelets contained decreased amounts of SNAP-23, which suggests the reduced secretion controlled by SNARE interactions. The melanosome biogenesis is blocked at the latest step in gunmetal (Nguyen, et al) (view diagram of melanosome blockage and melanosomal protein sorting here). Stinchcombe, et al reported that many of the secretory lysosomes from gm CTL fail to polarize and are left around the periphery of the cells (view diagram of lytic granule blockage in CTL cells here).
REFERENCE
- Detter JC, Zhang Q, Mules EH, Novak EK, Mishra VS, Li W, McMurtrie EB, Tchernev VT, Wallace MR, Seabra MC, Swank RT, Kingsmore SF. Rab geranylgeranyl transferase alpha mutation in the gunmetal mouse reduces Rab prenylation and platelet synthesis. Proc Natl Acad Sci U S A 2000 ; 97: 4144-9. PMID: 10737774
- McGarry MP, Reddington M, Novak EK, Swank RT. Survival and lung pathology of mouse models of Hermansky-Pudlak syndrome and Chediak-Higashi syndrome. Proc Soc Exp Biol Med. 1999; 220: 162-8. PMID: 10193444
- Nguyen T, Novak EK, Kermani M, Fluhr J, Peters LL, Swank RT, Wei ML. Melanosome morphologies in murine models of hermansky-pudlak syndrome reflect blocks in organelle development. J Invest Dermatol 2002; 119: 1156-64. PMID: 12445206
- Novak EK, Reddington M, Zhen L, Stenberg PE, Jackson CW, McGarry MP, Swank RT. Inherited thrombocytopenia caused by reduced platelet production in mice with the gunmetal pigment gene mutation. Blood 1995; 85: 1781-9. PMID: 7703484
- Richards-Smith B, Novak EK, Jang EK, He P, Haslam RJ, Castle D, Whiteheart SW, Swank RT. Analyses of proteins involved in vesicular trafficking in platelets of mouse models of Hermansky Pudlak syndrome. Mol Genet Metab 1999; 68: 14-23. PMID: 10479478
- Stinchcombe JC, Barral DC, Mules EH, Booth S, Hume AN, Machesky LM, Seabra MC, Griffiths GM. Rab27a is required for regulated secretion in cytotoxic T lymphocytes. J Cell Biol 2001; 152: 825-34. PMID: 11266472
- Swank RT, Jiang SY, Reddington M, Conway J, Stephenson D, McGarry MP, Novak EK. Inherited abnormalities in platelet organelles and platelet formation and associated altered expression of low molecular weight guanosine triphosphate-binding proteins in the mouse pigment mutant gunmetal. Blood 1993; 81: 2626-35. PMID: 8490171
- Zhang Q, Zhen L, Li W, Novak EK, Collinson LM, Jang EK, Haslam RJ, Elliott RW, Swank RT. Cell-specific abnormal prenylation of Rab proteins in platelets and melanocytes of the gunmetal mouse. Br J Haematol 2002; 117: 414-23. PMID: 11972527