GENOMIC
Mapping
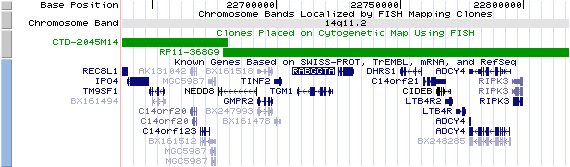
14q11.2 View the map and BAC clones of FISH (data from UCSC genome browser).
Structure
(assembly 07/03)
Isoform a (NM_182836): 17 exons, 6,058bp, Chr14: 22,724,874 - 22,730,931.
Isoform b (NM_004581): 16 exons, 6,058bp, Chr14: 22,724,874 - 22,730,931.
Note that NM_004581 differs from NM_182836 in the alternative splicing of exon 1, which leads to a shorter C-terminus. The exon/intron structural organization of the 5'-untranslated region (UTR) of human RABGGTA was found to be similar to that of the mouse Rabggta gene. 5'RACE shows variable 5' ends (Li, et al). van Bokhoven, et al found that the 3' end of the RABGGTA cDNA sequence overlaps the promoter region of TGM1.
The figure below shows the structure of the known isoforms (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
Features of the 5'-UTR of RABGGTA suggest it is a housekeeping gene (Li, et al).
TRANSCRIPT
RefSeq/ORF
a) Transcript variant 1 (NM_182836), 1,961bp, view ORF and the alignment to genomic.
a) Transcript variant 2 (NM_004581), 2,280bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: ubiquitously expressed.
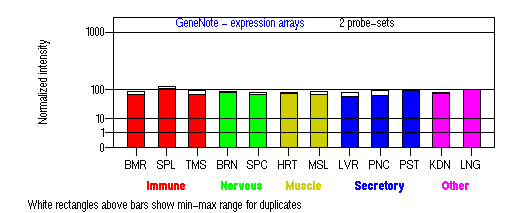
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle; LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
(Rab GGTase alpha)
Isoform a (NP_878256): 567aa, ExPaSy NiceProt view of Swiss-Prot:Q92696.
Isoform b (NP_004572): 297aa (encoded by transcript variant 2).
Synonyms: Geranylgeranyl transferase type II alpha subunit; Rab geranylgeranyltransferase alpha subunit; Rab GG transferase alpha.
Ortholog
| Species | Mouse | Rat | Zebrafish | Worm |
| GeneView | gm/Rabggta | LOC58983 | 19427 | M57.2 |
| Protein | NP_062392 (567aa) | NP_113842 (567aa) | 7197 (333aa) |
CE19541 (580aa) |
| Identities | 90%/515aa | 91%/518aa | 55%/186aa | 30%/182aa |
| Species | Mosquito | Fruitfly | Yeast | |
| GeneView | 1279131 | CG12007 | YJL031C | |
| Protein | XP_318802 (495aa) | Q9VN77 (515aa) |
YJL031C (327aa) | |
| Identities | 39%/190aa | 37%/176aa | 31%/86aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) Pfam:PPTA repeats:
47 - 77; 91 - 121; 127 - 157; 162 - 192; 210-240.
b) Pfam:LRR: 462 - 485, 507 - 533.
(2) Graphical view of InterPro domain structure.
(3) Transmembrane domains predicted by SOSUI: none.
(4) CDD domains:
a) COG5536: Protein prenyltransferase, alpha subunit (post-translational modification).
b) COG0529: Protein geranylgeranyltransferase type II, alpha subunit (post-translational modification).
Motif/Site
(1) Predicted results by ScanProsite:
a) Leucine-rich region profile : [occurs frequently]
385 - 559: score=10.339
b) Protein kinase C phosphorylation site : [occurs frequently]
78 - 80: TqK, 317 - 319: TfR, 370 - 372: ScK 410 - 412: TlK.
c) N-glycosylation site : [occurs frequently]
124 - 127: NWTR, 174 - 177: NFSN, 177 - 180: NYSS
d) Tyrosine kinase phosphorylation site : [occurs frequently]
393 - 400: RalDpllY
e) Leucine zipper pattern : [occurs frequently]
538 - 559: LnlqgnpLcqavgiLeqlaelL, 545 - 566: LcqavgiLeqlaelLpsvssvL
f) N-myristoylation site : [occurs frequently]
542 - 547: GNplCQ
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: XXRR-like motif in the N-terminus: HGRL
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
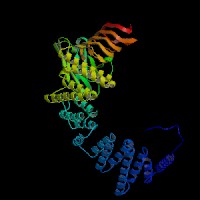
3D Model
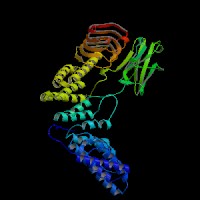
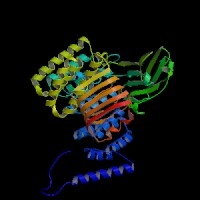
ModBase (Q92696): predicted comparative 3D structures (data from UCSC Genome Sorter).
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=65,072Da, pI=5.45 (NP_878256, isoform a).
Computed theoretical MW=33,767Da, pI=5.02 (NP_004572, isoform b).
FUNCTION
Ontology
a) Process: protein amino acid prenylation.
b) Function: protein prenyltransferase activity.
Location
Cytoplasmic.
Interaction
The RABGGTA gene encode the geranylgeranyl transferase type II alpha subunit (EC 2.5.1.60) (Rab geranylgeranyltransferase alpha subunit, or RabGGTase alpha). RabGGTase catalyzes the transfer of a geranyl-geranyl moiety from geranyl-geranyl pyrophosphate to both cysteines in Rab proteins with an -XXCC, -XCXC and -CCXX C-terminal, such as RAB1A, RAB3A, RAB5A, and RAB27A respectively. The enzymatic reaction requires the aid of a Rab escort protein (REP, also called component A). Heterodimer of an alpha and a beta subunit, is collectively called component B. The head-to-tail arrangement with the transglutaminase 1 (TGM1) gene does not show that these two genes are functionally related in the terminal differentiation program of epidermal keratinocytes (Song, et al).
2 proteins are shown to be associated with YJL031C/BET4 in Yeast GRID.
Rabggta drosophila homolog CG12007 interaction information in CuraGen interaction database.
Pathway
The enzyme RabGGTase covalently attaches the geranylgeranyl groups to two C-terminal cysteines of the Rab protein. NiceZyme View of protein geranylgeranyltransferase type II within ENZYME: EC 2.5.1.60. More details in BRENDA: RabGGTase.
MUTATION
Allele or SNP
Li, et al performed mutation screening of the human RABGGTA gene in 8 patients with deficiencies of platelet-dense granules (alphadelta-SPD), alpha granules (alpha-SPD or gray platelet syndrome, GPS) or alpha plus dense granules (alphadelta-SPD). Obvious disease-causing mutations of human RABGGTA were not found in these SPD patients by sequencing its entire coding region.
SNPs deposited in dbSNP.
Distribution
(none)
Effect
(none)
PHENOTYPE
(Animal Models)
Mutation in the Rabggta gene is the cause of gunmetal (gm) mutant (Detter, et al), a mouse model of Hermansky-Pudlak syndrome (OMIM 601905).
REFERENCE
- Detter JC, Zhang Q, Mules EH, Novak EK, Mishra VS, Li W, McMurtrie EB, Tchernev VT, Wallace MR, Seabra MC, Swank RT, Kingsmore SF. Rab geranylgeranyl transferase alpha mutation in the gunmetal mouse reduces Rab prenylation and platelet synthesis. Proc Natl Acad Sci U S A 2000 ; 97: 4144-9. PMID: 10737774
- Li W, Detter JC, Weiss HJ, Cramer EM, Zhang Q, Novak EK, Favier R, Kingsmore SF, Swank RT. 5'-UTR structural organization, transcript expression, and mutational analysis of the human Rab geranylgeranyl transferase alpha-subunit (RABGGTA) gene. Mol Genet Metab 2000; 71: 599-608. PMID: 11136552
- Song HJ, Rossi A, Ceci R, Kim IG, Anzano MA, Jang SI, De Laurenzi V, Steinert PM. The genes encoding geranylgeranyl transferase alpha-subunit and transglutaminase 1 are very closely linked but not functionally related in terminally differentiating keratinocytes. Biochem Biophys Res Commun 1997; 235: 10-4. PMID: 9196026
- van Bokhoven H, Rawson RB, Merkx GF, Cremers FP, Seabra MC. cDNA cloning and chromosomal localization of the genes encoding the alpha- and beta-subunits of human Rab geranylgeranyl transferase: the 3' end of the alpha-subunit gene overlaps with the transglutaminase 1 gene promoter. Genomics 1996; 38: 133-40. PMID: 8954794
EDIT HISTORY:
Created by Wei Li & Jonathan Bourne 07/06/2004