GENOMIC
Mapping
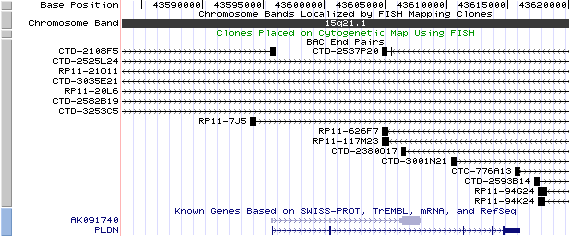
15q21.1. View the map and BAC clones (data from UCSC genome browser).
Structure
(assembly 07/03)
PLDN/NM_012388: 5 exons, 22,492bp, Chr15: 43,595,473-43,617,964.
Transcript 1 contains five coding exons (AF080470), and transcript 2 has three coding exons (AK128626); only
exon 2 is shared by both transcripts.
The figure below shows the map of the PLDN gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
PLDN (NM_012388), 3,959bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: Ubiquitously expressed of transcript 1 , with the highest expression levels observed in brain, heart, liver and kidney. In contrast, transcript 2 was expressed only in adult brain, testis, and leukocytes and in fetal brain, lung, and thymus. Transcript 2 is not expressed in normal fibroblasts or melanocytes (Cullinane, et al).
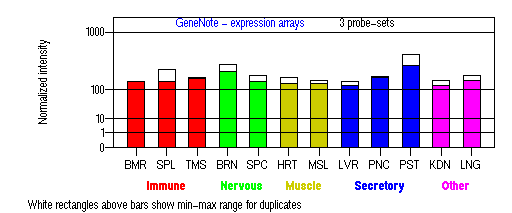
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle; LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
Pallidin (NP_036520): 172aa, ExPaSy NiceProt view of Swiss-Prot:Q9UL45.
Synonyms: pallid (mouse) homolog, pallidin; syntaxin 13-interacting
protein pallid; syntaxin 13 binding protein 1.
Ortholog
| Species | Mouse | Chimp | Rat | Drosophila |
| GeneView | Pallid | 07038 | LOC317630 | CG14133 |
| Protein | NP_036520 (172aa) | 12031(172aa) | XP_229231(692aa) | NP_648494(120aa) |
| Identities | 87%/149aa | 100%/172aa | 86%/148aa | 32%/36aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) coiled coil: 68 - 89.
b) coiled coil: 125 - 167.
(2) Transmembrane domains predicted by SOSUI: None.
Motif/Site
(1) Predicted results by ScanProsite:
a) Casein kinase II phosphorylation site : [occurs frequently]
7 - 10: SspD,
32 - 35: TspD,
33 - 36: SpdE,
43 - 46: TieD,
97 - 100: SmlD.
b) N-myristoylation site : [occurs frequently]
28 - 33: GLsdTS.
c) Protein kinase C phosphorylation site : [occurs frequently]
129 - 131: TsK,
165 - 167: TaR.
d) Bipartite nuclear targeting sequence : [occurs frequently]
119 - 135:
RKemlmlhektsklkkr.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: found KKXX-like motif in the C-terminus: PAKR
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: None.
3D Model
(1) ModBase No entry found.
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
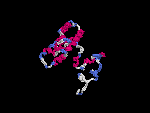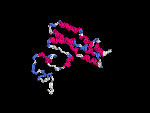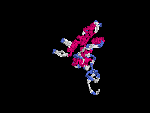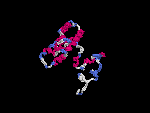

2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=19,744Da, pI=6.01 (NP_036520).
FUNCTION
Ontology
(1) Process: pigmentation, vesicle-mediated transport.
(2) Protein interaction in BLOC-1.
(3) Synaptic vesicle docking.
Location
Cytoplasmic. It can be a membrane associated protein.
Interaction
Pallidin interacts with muted, cno, dysbindin, Blos1, syntaxin 13, and F-actin (Ciciotte, et al; Falcon-Perez, et al; Li, et al (2004); Starcevic, et al). Pallidin is a subunit of the biogenesis of lysosome-related organelles complex 1 (BLOC-1), which contains the products of seven other HPS genes, sdy, mu, cno, rp, Snapap, Blos1, Blos2 (Ciciotte, et al; Falcon-Perez , et al; Li, et al (2003); Moriyama, et al; Starcevic, et al) (view diagram of BLOC-1 complex here). Syntaxin-13 is a member of t-SNARE which is localized to the endosomal membrane and is required for vesicle fusion to the early enddosomes and recycling endosomes. Yeast two-hybrid analyses revealed that pallidin is capable of self-association through a region that contains its two coiled-coil forming domains. Immunofluorescence and in vitro binding experiments demonstrated that pallidin/BLOC-1 is able to associate with actin filaments ( Falcon-Perez, et al ). More details about the function of BLOC-1 are described in the HPS7 profile.
Pallidin drosophila homolog CG14133 interaction information in CuraGen interaction database.
Pathway
Involved in the development of lysosome-related organelles, such as melanosomes and platelet-dense granules (view diagram of BLOC-1 pathway here).
MUTATION
Allele or SNP
SNPs deposited in dbSNP.
Distribution
| Location | Genomic | cDNA | Protein | Type | Ethnicity | Reference |
| Exon 3 | 232C>T | 232C>T | Q78X | nonsense | Indian | Cullinane, et al |
Effect
The Q78X mutation does not cause nonsense mediated decay (NMD) directly as the transcript with predicted size is obvious. Exonic Splicing Enhancers (ESE) finder tool indicated that c.232C>T would abolish a SC35 binding site in the DNA; abolishment of this binding site is predicted to result in the skipping of exon3. Indeed, sequencing of the patient's additional smaller PCR band revealed the absence of exon 3 (88 bp) and puts exon 4 out of frame, which might introduce susceptibility to nonsense-mediated decay or create an alternate protein consisting of 86 amino acids: 74 of the original pallidin N-terminal amino acids encoded by exons 1 and 2 and then 12 alternate amino acids encoded by exon 4 (Cullinane, et al).
PHENOTYPE
The HPS-9 patient exhibit typical HPS symptoms including oculocutaneous albinism with nystagmus, iris transillumination and retinal hypopigmentation, and absent platelet delta granules. However, there is no history of severe or unusual infections, easy bruising, or bleeding. The melanogenic protein TYRP1 showed aberrant localization, an increase in plasma-membrane trafficking, and a failure to reach melanosomes (Cullinane, et al).
(Animal Models)
Mutation in the Pldn gene is the cause of pallid (pa) mutant (Huang, et al), a mouse model of Hermansky-Pudlak syndrome (OMIM 604310).
REFERENCE
- Ciciotte SL, Gwynn B, Moriyama K, Huizing M, Gahl WA, Bonifacino JS, Peters LL. Cappuccino, a mouse model of Hermansky-Pudlak syndrome, encodes a novel protein that is part of the pallidin-muted complex (BLOC-1). Blood 2003; 101: 4402-7. PMID: 12576321
- Cullinane AR, Curry JA, Carmona-Rivera C, Summers CG, Ciccone C, Cardillo ND, Dorward H, Hess RA, White JG, Adams D, Huizing M, Gahl WA. A BLOC-1 Mutation Screen Reveals that PLDN Is Mutated in Hermansky-Pudlak Syndrome Type 9. Am J Hum Genet 2011; 88: 778-87. PMID: 21665000
- Falcon-Perez JM, Starcevic M, Gautam R, Dell'Angelica EC. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem 2002; 277: 28191-9. PMID: 12019270
- Huang L, Kuo YM, Gitschier J. The pallid gene encodes a novel, syntaxin 13-interacting protein involved in platelet storage pool deficiency. Nat Genet 1999; 23: 329-32. PMID: 10610180
- Li W, Rusiniak ME, Chintala S, Gautam R, Novak EK, Swank RT. Murine Hermansky-Pudlak syndrome genes: regulators of lysosome-related organelles. Bioessays 2004; 26: 616-28. PMID: 15170859
- Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O'Brien EP, Tinsley CL, Blake DJ, Spritz RA, Copeland NG, Jenkins NA, Amato D, Roe BA, Starcevic M, Dell'Angelica EC, Elliott RW, Mishra V, Kingsmore SF, Paylor RE, Swank RT. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat Genet 2003; 35: 84-9. PMID: 12923531
- Moriyama K, Bonifacino JS. Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic 2002; 3: 666-77. PMID: 12191018
- Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J Biol Chem 2004; 279: 28393-401. PMID: 15102850
EDIT HISTORY:
Created by Wei Li & Jonathan Bourne 07/08/2004
Updated by Wei Li, 07/24/2011
Updated by Wei Li, 08/03/2012
Updated by Wei Li, 06/13/2013