GENOMIC
Mapping
1p31.3. View the map and BAC clones (data from UCSC genome browser).
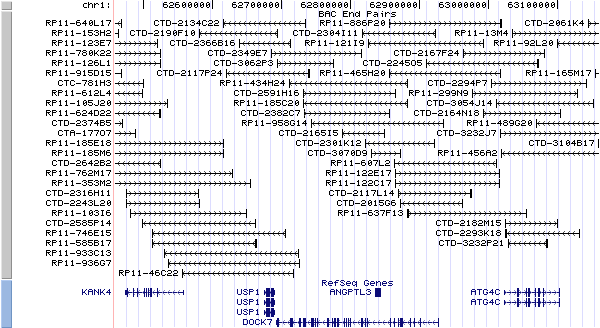
Structure
(assembly 03/2006)
DOCK7 (NM_033407): 49 exons, 233,573 bp, chr1:62,692,985-62,926,557.
The figure below shows the structure of the DOCK7 gene (data from UCSC genome browser).
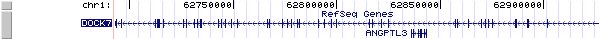
Regulatory Element
TRANSCRIPT
RefSeq/ORF
DOCK7 (NM_033407), 7,063 bp, view ORF and the alignment to genomic.
Expression Pattern
Widely expressed. Highly expressed in the brain. View more information in Entrez Gene, or UCSC Gene Sorter, or GeneCards.
PROTEIN
Sequence
DOCK7 (Dedicator of cytokinesis protein 7, KIAA1771) (NP_212132): 2109 aa, 6 isoforms have been reported. UniProtKB/Swiss-Prot entry Q98N67.
Ortholog
View NCBI HomoloGene of DOCK7.
Domain
Domain: Pfam:PF06920.
Two domains are shared amongst all DOCK proteins, the catalytic DHR-2 (DOCK homology region 2) or CZH-2 (CDM-zizimin homology 2) domain and the DHR-1 or CZH-1domain. For all DOCK proteins, the DHR-1 domain is located N-terminal to the DHR-2 domain. The DHR-1 domain is probably involved in the binding with PtdIns, while the DHR-2 domain may be responsible for the GEF activity through the interaction with Rho GTPase.
Motif/Site
View annotated motifs in UniProt.
3D Model
ModBase predicted 3D structure of Q96N67 from UCSC Gene Sorter: (none).
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=239,422Da, pI=6.34.
View NCBI HomoloGene of DOCK7.
Domain
Domain: Pfam:PF06920.
Two domains are shared amongst all DOCK proteins, the catalytic DHR-2 (DOCK homology region 2) or CZH-2 (CDM-zizimin homology 2) domain and the DHR-1 or CZH-1domain. For all DOCK proteins, the DHR-1 domain is located N-terminal to the DHR-2 domain. The DHR-1 domain is probably involved in the binding with PtdIns, while the DHR-2 domain may be responsible for the GEF activity through the interaction with Rho GTPase.
Motif/Site
View annotated motifs in UniProt.
3D Model
ModBase predicted 3D structure of Q96N67 from UCSC Gene Sorter: (none).
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=239,422Da, pI=6.34.
FUNCTION
Ontology
(1) Biological process: differentiation, neurogenesis.
(2) Guanine-nucleotide releasing factor, guanyl-nucleotide exchange factor activity.
(3) GTP binding, Rac GTPase binding.
(4) Regulation of pigmentation during development.
Location
Cell projection.
Interaction
Interacts with TSC1. Interacts with nucleotide-free RAC1 and RAC3.
View interactions in HPRD
View co-occured partners in literature searched by PPI Finder.
Pathway
Functions as a guanine nucleotide exchange factor (GEF), which activates Rac and Cdc42 small GTPases by exchanging bound GDP for free GTP. Does not have a GEF activity for RhoA. Required for STMN1 'Ser-15' phosphorylation during axon formation and consequently for neuronal polarization.
PHENOTYPE
Mutaitons of Dock7 in mice cause the phenotype of white belly spots in misty(m) and moonlight (mnlt) (Blasius et al.). More details has been described in Mutagenetix.
REFERENCE
- Blasius AL, Brandl K, Crozat K, Xia Y, Khovananth K, Krebs P, Smart NG, Zampolli A, Ruggeri ZM, Beutler BA. Mice with mutations of Dock7 have generalized hypopigmentation and white-spotting but show normal neurological function. Proc Natl Acad Sci U S A 2009; 106:2706-11. PMID: 19202056