GENOMIC
Mapping
7qA3. View the map and BAC contig (data from UCSC genome browser).
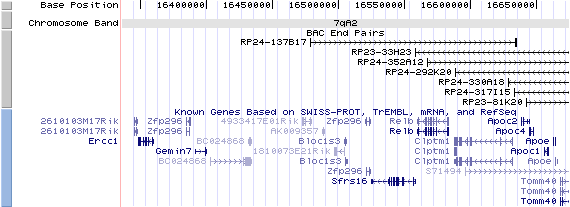
Structure
(assembly 10/03)
Bloc1s3/NM_177692: 2 exons, 1,883 bp, chr7:16,504,517-16,506,399.
The figure below shows the structure of the Bloc1s3 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=human BLOC1S3, seq2=mouse rp) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Bloc1s3/NM_177692: 1,189 bp, view ORF and the alignment to genomic.
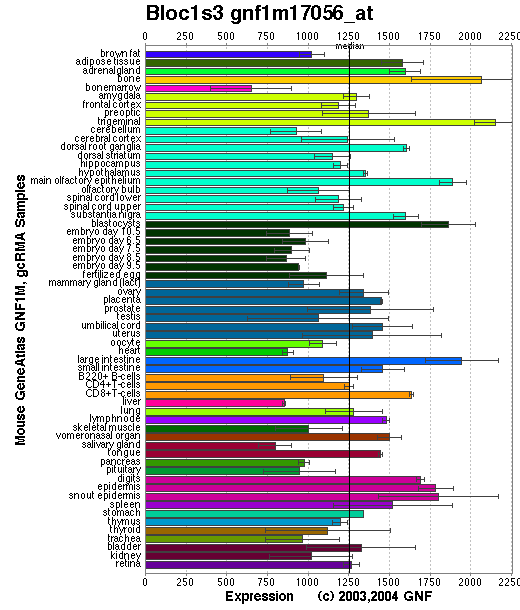
Expression Pattern
Tissue specificity: Ubiquitously expressed (by similarity).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
BLOC-1 subunit 3
(NP_808360): 195aa. ExPaSy NiceProt view of Swiss-Prot:Q8C6R4.
Synonyms: Hps8 protein, reduced pigmentation protein, E230011O18 product: hypothetic protein.
Ortholog
| Species | Human | Rat |
| GeneView | BLOC1S3 | LOC308413 |
| Protein | NP_997715 (202aa) | XP_218422 (195aa) |
| Identities | 87%/201aa | 93%/195aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains of predicted by SMART:
a) low complexity: 18 - 51
b) low complexity: 55 - 79
c) low complexity: 126 - 150
(2) Transmembrane domains predicted by SOSUI: none.
Motif/Site
(1) Predicted results by ScanProsite:
a) Amidation site : [occurs frequently]
5 - 8: qGRR.
b) Casein kinase II phosphorylation site : [occurs frequently]
22 - 25: TdsE,
29 - 32: SssE,
30 - 33: SseE,
31 - 34: SeeE,
59 - 62: TdsE,
61 - 64: SepE,
111 - 114: TrlD.
c) N-myristoylation site : [occurs frequently]
49 - 54: GLrvAG,
118 - 123: XAavSG,
146 - 151: GLaaAH.
d) Protein kinase C phosphorylation site : [occurs frequently]
152 - 154: SvR.
e) Cell attachment sequence : [occurs frequently]
157 - 159: RGD.
f) Tyrosine sulfation site : [occurs frequently]
29 - 43:
ssseeelYlgpsgpt.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER membrane retention signals: none
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase: none.
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
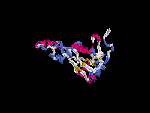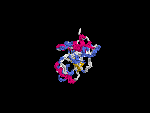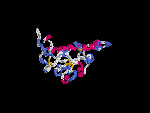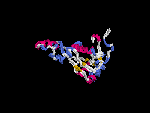
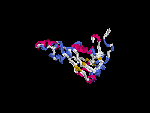
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=20,445Da, pI=4.84.
The phosphorylated form migrates slower than the unphosphorylated form (Gwynn, et al).
FUNCTION
Ontology
(1) May play a role in intracellular vesicle trafficking.
(2) Protein interaction in BLOC-1.
Location
Cytoplasmic.
Interaction
BLOC-1 subunit 3 is a subunit of the biogenesis of lysosome-related organelles complex 1 (BLOC-1), which resides with the products of seven other HPS genes, sdy, mu, pa, cno, Snapap, Blos1, Blos2 (Ciciotte, et al; Falcon-Perez , et al; Li, et al; Moriyama, et al; Starcevic, et al). It interacts with Blos2 within the complex ( Starcevic, et al) (view diagram of BLOC-1 complex here). More details about the function of BLOC-1 are described in the HPS7 profile.
Pathway
Involved in the development of lysosome-related organelles, such as melanosomes and platelet-dense granules (view diagram of BLOC-1 pathway here). Nguyen, et al found that the maturation of melanosomes is blocked in early stages (view diagram of melanosome blockage here).
MUTATION
Allele or SNP
1 phenotypic allele is described in MGI:2678952.
SNPs deposited in dbSNP.
Distribution
| Location | Genomic | cDNA | Protein | Type | Strain | Reference |
| Exon 2 | 238C>T | 238C>T | Q80X | nonsense | rp (B6) | Starcevic, et al |
Effect
The Q80X mutation is not subject to nonsense mediated decay, but no detectable Blos3 protein in liver extracts of rp mutants. The mutation does affect the stability of other subunits such as pallidin and muted of BLOC-1 complex, but does not completely disrupt BLOC-1 assembly (Starcevic, et al).