GENOMIC
Mapping
3qF1. View the map and BAC contig (data from UCSC genome browser).
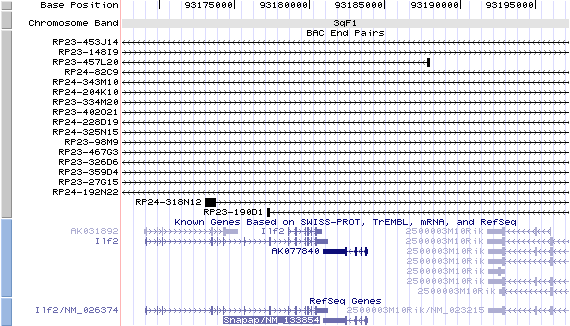
Structure
(assembly 10/03)
Snapap/NM_133854: 4 exons, 2,971 bp, chr3:93,180,903-93,183,873.
Note that the 3'-UTRs of both Snapap and Ilf2 gene overlap.
The figure below shows the structure of the Snapap gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=human SNAPAP, seq2=mouse Snapap) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Snapap/NM_133854: 1,898 bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: Ubiquitously expressed. Strongly expressed in heart, brain, testis, kidney and liver; low expression in spleen, lung and skeletal muscle.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
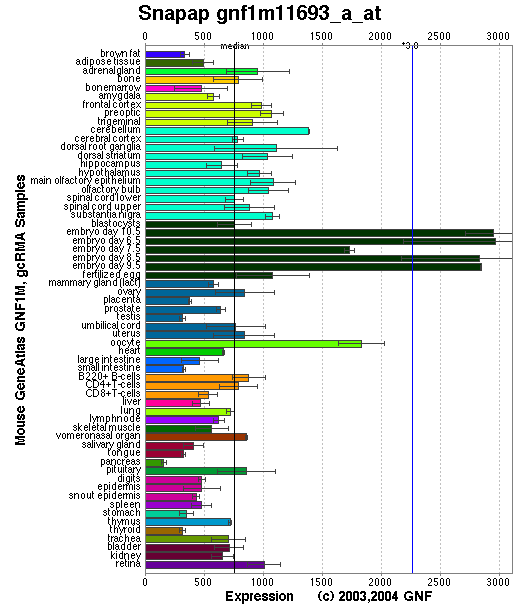
Tissue specificity: Ubiquitously expressed. Strongly expressed in heart, brain, testis, kidney and liver; low expression in spleen, lung and skeletal muscle.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.

PROTEIN
Sequence
Snapin
(NP_598615): 136aa, ExPaSy NiceProt view of Swiss-Prot:Q9Z266.
Synonyms: Synaptosomal-associated protein 25 binding protein (Snap25bp), SNARE-associated protein Snapin, SNAP-associated protein.
Ortholog
| Species | Human | Rat | Zebrafish | Drosophila | Worm |
| GeneView | SNAPAP | LOC295217 | 02356 | CG32951-PA | C02B10.2 |
| Protein | NP_036569 (136aa) | XP_215602 (136aa) | 21677 (119aa) | NP_787967 (134aa) | WP:CE24776 (122aa) |
| Identities | 97%/136aa | 100%/136aa | 83%/119aa | 38%/107aa | 31%/93aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) low complexity: 2 - 19
b) coiled coil: 44-123
(2) Transmembrane domains predicted by SOSUI: none.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-myristoylation site : [occurs frequently]
13 - 18: GTpvAG.
b) Protein kinase C phosphorylation site : [occurs frequently]
20 - 22: TgR,
117 - 119: TaR.
c) Casein kinase II phosphorylation site : [occurs frequently]
20 - 23: TgrD,
50 - 53: SqvE.
PKA-phosphorylation of Ser50 increased binding of synaptagmin to SNARE complex (Chheda, et al).
d) Tyrosine kinase phosphorylation site : [occurs frequently]
73 - 81: KvalDldpY,
121 - 129: RamlDsgvY.
e) N-glycosylation site : [occurs frequently]
110 - 113: NHSV.
f) Bipartite nuclear targeting sequence : [occurs frequently]
107 - 123:
RRlnhsvaketarrram.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER membrane retention signals: none
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase: none.
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
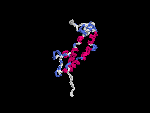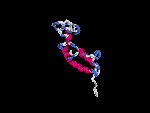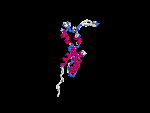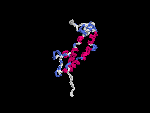
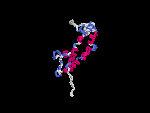
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=14,904Da, pI=9.35.
FUNCTION
Ontology
(1) Process: synaptic vesicle exocytosis.
(2) Component of synaptosome.
(3) SNARE binding.
(4) May play a role in intracellular vesicle trafficking.
(2) Protein interaction in BLOC-1.
Location
May be cytoplasmic and peripheral membrane bound or anchored to the vesicular membrane through an N- terminal signal anchor. Ilardi, et al found that snapin is exclusively associated with the snaptic vesicle fraction and absent from the cytosolic and plasma membrane fractions. They concluded snapin behaves as an integral, vesicle-associated, membrane protein. Buxton, et al reported that 70% of total cellular snapin is present in the cytosol.
Interaction
Snapin is a subunit of the biogenesis of lysosome-related organelles complex 1 (BLOC-1), which contains the products of seven other HPS genes, sdy, mu, pa, cno, rp, Blos1, Blos2 (Ciciotte, et al; Falcon-Perez , et al; Li, et al; Moriyama, et al; Starcevic, et al). It interacts with dysbindin, cno, Blos1, and Blos2 within the complex ( Starcevic, et al) (view diagram of BLOC-1 complex here). The same 69-residue region of dysbindin that is sufficient for dystrobrevin binding in vitro also contains the binding sites for pallidin and snapin, and at least part of the muted-binding interface (Nazarian, et al). Feng, et al further narrowed down the interaction domain to dysbindin 90-119 peptide. Snapin is a binding partner of dysbindin-1 in vitro and in the brain. Tissue fractionation of whole mouse brains and human hippocampal formations revealed that both dysbindin-1 and snapin are concentrated in tissue enriched in synaptic vesicle membranes and less commonly in postsynaptic densities. It is not detected in presynaptic tissue fractions lacking synaptic vesicles (Talbot, et al). Loss of dysbindin reduces 1/4 of snapin in hippocampal tissues which may affect neuronal transmission efficacy that leads to the development of schizophrenic symptoms (Feng, et al).
Snapin may associate with the SNARE complex. The interactions with SNAP23 and SNAP25 are controversial. SNAP23 is a ubiquitously expressed SNAP25-related protein. Ilardi, et al firstly identified the snapin by using SNAP25 as a bait to screen a human brain cDNA library. Snapin bound to GST-SNAP25 at a stoichiometry of 1:1. No binding was detectable to SNAP23, VAMP2, and syntaxin 1A. The C-terminal half of snapin was sufficient for the interaction with SNAP25. Snapin was a SNAP25 binding protein that forms a complex by association with syntaxin, SNAP25 and VAMP2. Snapin was required for high-affinity synaptotagmin-SNARE interation. PKA-phosphorylation of Ser50 increased its interaction with SNAP25 (Chheda, et al). Overexpression of Snapin S50D, a mutant mimicking the phosphorylated state, resulted in a decreased number of readily releasable vesicles (Thakur, et al). Expression of Snapin-C66A, a dimerization-defective mutant with impaired interactions with SNAP-25 and Synaptotagmin, reduces the readily releasable pool size but exhibits less effect on synchronized fusion (Pan, et al).
In vitro and in vivo protein-protein interaction assays showed that snapin interacted with SNAP23 and the C-terminal helical domain of snapin contained the SNAP-23-binding site. Snapin formed a complex with SNAP23 and syntaxin 4, but snapin did not affect the affinity of syntaxin 4 and SNAP23 (Buxton, et al). In pull-down studies, snapin detected AQP2, syntaxin-3, syntaxin-4, and SNAP23 from the inner medullary collecting duct. Snapin plays an important role as a linker between the water channel and the t-SNARE complex, the association of AQP2 with both syntaxin-3 and syntaxin-4 is highly enhanced by the presence of snapin (Mistry, et al (2009)). Vites, et al were unable to detect any specific interaction between snapin and SNAP25 (or SNAP23) by pulldown experiments, co-IP, CD- and fluorescence anisotropy spectroscopy. Recombinant snapin forms a stable dimmer with a predominantly alpha-helical secondary structure. Snapin is required to maintain a proper balance of the late endocytic protein LAMP-1 and late endosomal SNARE proteins syntaxin 8 and Vti1b. Deleting the snapin gene in mice selectively led to the accumulation of these proteins in late endocytic organelles (Lu, et al). Snapin regulates the recruitment of late endosomes to the dynein motor complex for retrograde trafficking along microtubules and maturation of lysosomes (Cai, et al).
Except for the interaction between snapin and SNARE complex, several snapin binding partners have been found.
(1) Hunt, et al identified that snapin interacts with the N-terminus of regulator of G protein signaling 7 (RGS7) by using yeast two-hybrid analysis to screen a human whole brain cDNA library. The interaction is mediated primarily by amino acid residues 1-69 of RGS7 (which contains the proximal portion of the DEP domain).
(2) Snapin interacts with type VI adenylyl cyclase (ACVI) via amino acids 1 approximately 86 of ACVI and 33-51 of Snapin. Phosphorylation of Snapin by PKC or PKA might not be crucial for Snapin action on ACVI (Chou, et al).
(3) Receptor tyrosine kinase MET is a putative interacting protein of snapin (Schaaf, et al).
(4) The estrogen receptor-binding fragment-associated gene9 (EBAG9) gene product was recently identified as a modulator of tumor-associated O-linked glycan expression in nonneuronal cells. EBAG9 interacts with Snapin, which is likely to be a modulator of Synaptotagmin-associated regulated exocytosis. EBAG9 decreased phosphorylation of Snapin, which has a regulatory function in exocytosis (Ruder, et al).
(5) The vanilloid receptor-1 (TRPV1) plays a key role in the perception of peripheral thermal and inflammatory pain. Snapin and synaptotagmin IX (Syt IX), strongly interact in vitro and in vivo with the TRPV1 N-terminal domain. PKC signaling promotes at least in part the SNARE-dependent exocytosis of TRPV1 to the cell surface (Morenilla-Palao, et al).
(6) Using a yeast two-hybrid screen of a rat brain cDNA library with cypin lacking the carboxyl terminal eight amino acids as bait, snapin was identified as a cypin binding partner. The carboxyl-terminal coiled-coil domain (H2) of snapin is required for cypin binding. Snapin competes with tubulin for binding to cypin through cypin's CRMP homology domain, resulting in decreased microtubule assembly, therefore snapin regulates dendrite number in developing neurons (Chen, et al).
(7) Collectrin binds to SNARE complexes by interacting with snapin and facilitated SNARE complex formation. Therefore, collectrin is a regulator of SNARE complex function, which thereby controls insulin exocytosis (Fukui, et al).
(8) The interaction of granulocyte colony-stimulating factor (GCSF-R) and Snapin was found by yeast two-hybrid experiment by screen a mouse liver library. This interactionwas further confirmed by GST pull-down experiment, mammalian two-hybrid experiment and co-immunoprecipitation study. The immuno-fluorescence assay was shown that the two proteins of GCSF-R with Snapin were co-localized in the cytoplasm and plasma membrane. The region of C-terminal GCSF-R between box2 and box3, including the residue Tyr703, was responsible for the interaction with Snapin (Yuan, et al).
(9) Snapin binds to a 170-residue predicted ryanodine receptor (RyR) cytosolic loop (RyR2 residues 4596-4765), containing a hydrophobic segment required for snapin interaction. Deletion analysis indicates that the ryanodine receptor interacts with the snapin C-terminus, the same region as the SNAP25-binding site. Competition experiments with native ryanodine receptor and SNAP25 suggest that these two proteins share an overlapping binding site on snapin. The snapin-RyR1 association appears to sensitise the channel to Ca(2+) activation in [(3)H]ryanodine-binding studies (Zissimopoulos, et al).
(10) TRPM7, a member of the transient receptor potential (TRP) ion channel family, resides in the membrane of synaptic vesicles of sympathetic neurons, forms molecular complexes with the synaptic vesicle proteins synapsin I and synaptotagmin I, and directly interacts with synaptic vesicular snapin. Changes in TRPM7 levels and channel activity alter acetylcholine release, as measured by EPSP amplitudes and decay times in postsynaptic neurons. TRPM7 affects EPSP quantal size, and targeted peptide interference of TRPM7's interaction with snapin affects the amplitudes and kinetics of postsynaptic EPSPs (Krapivinsky, et al).
(11) Wolff, et al identified snapin as an interaction partner of the CK1 isoform delta (CK1delta) in the yeast two-hybrid system and confirmed the interaction by co-immunoprecipitation. Snapin was phosphorylated by CK1delta in vitro. Both proteins localized in close proximity in the perinuclear region, wherein snapin was found to associate with membranes of the Golgi apparatus.
(12) Suzuki, et al identified snapin as a protein interacting with the C terminus of the alpha(1A)-AR (alpha(1A)-adrenoceptor), and found that snapin co-immunoprecipitated with both TRPC6 (transient receptor potential canonical channel 6) and alpha(1A)-AR, and these interactions were augmented upon alpha(1A)-AR activation, increasing the recruitment of TRPC6 to the cell surface. In receptor-expressing PC12 cells, co-transfection of Snapin augmented alpha(1A)-AR-stimulated sustained increases in intracellular Ca(2+) ([Ca(2+)](i)) via ROC channels.
(13) The C-terminal coiled-coil domain (H2) of snapin is required for UT-A1 (urea transporter A1) interaction. Snapin binds to the intracellular loop of UT-A1 but not to the N- or C-terminal fragments. GST-pulldown experiments and co-immunoprecipitation studies verified that snapin interacts with native UT-A1, SNAP23, and syntaxin-4 in rat kidney inner medulla. Confocal analysis shows UT-A1 and snapin co-localised in both the cytoplasm and in the plasma membrane (Mistry, et al (2007)).
(14) The interaction between Exo70 and Snapin is mediated via an N-terminal coil-coil domain in Exo70 and a C-terminal helical region in Snapin. Exo70 competes with SNAP23 for Snapin binding, suggesting that Snapin does not provide a direct link between the exocyst and the SNARE complex but, rather, mediates cross-talk between the two complexes by sequential interactions. Depletion of Snapin in adipocytes using RNA interference inhibits insulin-stimulated glucose uptake, suggesting Snapin interacts with the exocyst and plays a modulatory role in GLUT4 vesicle trafficking (Bao, et al). Snapin mediates incretin action and augments glucose-dependent insulin secretion (Song, et al).
(15) An AAA-ATPase torsinA (TA) is an interacting partner of Snapin. Snapin co-localised with endogenous torsinA on dense core granules in PC12 cells in regulating exocytosis (Granata, et al (2008)). TA binds CSN4 and snapin in neuroblastoma cells and in brain synaptosomes. CSN4 and TA are required for the stability of both snapin and the synaptotagmin-specific endocytic adaptor stonin 2. Snapin is phosphorylated by the CSN-associated kinase protein kinase D (PKD) and its expression is decreased upon PKD inhibition. (Granata, et al (2011))
(16) PUMILIO2 forms a complex with NANOS1 in human male germ cells. In addtion, both interacts with snapin (Ginter-Matuszewska, et al).
(17) Yeast two hybrid screening experiments showed that EHD1 interacts with snapin. EHD1 binds to the C terminus of snapin via its C terminus EH domain. It negatively affects the binding of a SNARE complex protein, SNAP-25, to snapin, probably due to the competition for overlapping binding sites on the C terminus of snapin. EHD1 affects the coupling of synaptotagmin-1 to the SNARE complex, and could be a negative regulator of exocytosis. (Wei, et al).
(18) TorsinA (TA) binds CSN4 and snapin in neuroblastoma cells and in brain synaptosomes. CSN4 and TA are required for the stability of both snapin and the synaptotagmin-specific endocytic adaptor stonin 2. Snapin is phosphorylated by the CSN-associated kinase protein kinase D (PKD) and its expression is decreased upon PKD inhibition. (Granata, et al).
(19) Ca(2+) entry via the alpha(1A)-AR-Snapin-TRPC6-pathway plays an important role in physiological regulation of cardiac contractility. This pathway involves alpha(1A)-AR-activated translocation of Snapin and the transient receptor potential canonical 6 (TRPC6) channel to the plasma membrane (Mohl, et al
Snapin drosophila homolog CG32951 interaction information in CuraGen interaction database.
Pathway
Snapin is a component of the SNARE complex of proteins that is required for synaptic vesicle docking and fusion. It may modulate a step between vesicle priming, fusion and calcium-dependent neurotransmitter release by potentiating the interaction of synaptotagmins with the SNAREs and the plasma- membrane-associated protein SNAP25 (Ilardi, et al). It may also have a role in the mechanisms of SNARE-mediated membrane fusion in non-neuronal cells.
Snapin may be involved in the development of lysosome-related organelles, such as melanosomes and platelet-dense granules (view diagram of BLOC-1 pathway here).
MUTATION
llele or SNP
No phenotypic allele is described in MGI:1333745.
No mutation has been reported in mouse mutants.
SNPs deposited in dbSNP.
Distribution
(none)
Effect
(none)
PHENOTYPE
Snapin knock-out (KO) mice were produced via homologous recombination. In the snapin KO mice, the association of synaptagmin-1 with SNARE complex is decreased. The absence of snapin in embryonic chromaffin cells leads to a significant reduction of calcium-dependent exocytosis resulting from a decreased number of vesicles in releasable pools rather than abnormal biogenesis or morphological docking of large dense-core vesicles (LDCVs). This suggests that snapin plays a critical role in modulating neurosecretion by stabilizing the release-ready vesicles and modulating vesicle priming (Tian, et al). In addition to reduced frequency of miniature excitatory postsynaptic currents (mini-EPSCs) and smaller release-ready vesicle pool size, snapin deficiency results in EPSCs with multiple peaks and increased rise and decay times, reflecting "desynchronized" SV fusion (Pan, et al). Snapin plays a critical role in co-ordinating dynein-driven retrograde transport and late endosomal-lysosomal trafficking, thus maintaining efficient autophagy-lysosomal function (Cai, et al). Embryonic snapin-/- mouse brain showed reduced cortical plates and intermediate zone cell density, increased apoptotic death in the cortex and third ventricle, enhanced membrane-bound LC3-II staining associated with autophagic vacuoles and an accumulation of polyubiquitinated proteins in the cortex and hippocampus (Zhou, et al).
REFERENCE
- Bao Y, Lopez JA, James DE, Hunziker W. Snapin interacts with the Exo70 subunit of the exocyst and modulates GLUT4 trafficking. J Biol Chem 2008; 283: 324-31.PMID: 17947242
- Buxton P, Zhang XM, Walsh B, Sriratana A, Schenberg I, Manickam E, Rowe T. Identification and characterization of Snapin as a ubiquitously expressed SNARE-binding protein that interacts with SNAP23 in non-neuronal cells. Biochem J 2003; 375: 433-40. PMID: 12877659
- Cai Q, Lu L, Tian JH, Zhu YB, Qiao H, Sheng ZH. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron 2010; 68:73-86. PMID: 20920792
- Chen M, Lucas KG, Akum BF, Balasingam G, Stawicki TM, Provost JM, Riefler GM, Jornsten RJ, Firestein BL. A novel role for snapin in dendrite patterning: interaction with cypin. Mol Biol Cell,2005; 16: 5103-14. PMID: 16120643
- Chheda MG, Ashery U, Thakur P, Rettig J, Sheng ZH. Phosphorylation of Snapin by PKA modulates its interaction with the SNARE complex. Nat Cell Biol 2001; 3: 331-8. PMID: 11283605
- Chou JL, Huang CL, Lai HL, Hung AC, Chien CL, Kao YY, Chern Y. Regulation of type VI adenylyl cyclase by Snapin, a SNAP25-binding protein. J Biol Chem 2004; 279: 46271-9. PMID: 15319443
- Ciciotte SL, Gwynn B, Moriyama K, Huizing M, Gahl WA, Bonifacino JS, Peters LL. Cappuccino, a mouse model of Hermansky-Pudlak syndrome, encodes a novel protein that is part of the pallidin-muted complex (BLOC-1). Blood 2003; 101: 4402-7. PMID: 12576321
- Falcon-Perez JM, Starcevic M, Gautam R, Dell'Angelica EC. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem 2002; 277: 28191-9. PMID: 12019270
- Feng YQ, Zhou ZY, He X, Wang H, Guo XL, Hao CJ, Guo Y, Zhen XC, Li W. Dysbindin deficiency in sandy mice causes reduction of snapin and displays behaviors related to schizophrenia. Schizophr Res 2008; 106: 218-28. PMID: 18774265
- Fukui K, Yang Q, Cao Y, Takahashi N, Hatakeyama H, Wang H, Wada J, Zhang Y, Marselli L, Nammo T, Yoneda K, Onishi M, Higashiyama S, Matsuzawa Y, Gonzalez FJ, Weir GC, Kasai H, Shimomura I, Miyagawa J, Wollheim CB, Yamagata K. The HNF-1 target Collectrin controls insulin exocytosis by SNARE complex formation. Cell Metab 2005; 2:373-84. PMID: 16330323
- Ginter-Matuszewska B, Spik A, Rembiszewska A, Koyias C, Kupryjanczyk J, Jaruzelska J. The SNARE-associated component SNAPIN binds PUMILIO2 and NANOS1 proteins in human male germ cells. Mol Hum Reprod 2009; 15: 173-9. PMID: 19168546
- Granata A, Watson R, Collinson LM, Schiavo G, Warner TT. The dystonia-associated protein torsina modulates synaptic vesicle recycling. J Biol Chem 2007; [Epub ahead of print] PMID: 18167355
- Granata A, Koo SJ, Haucke V, Schiavo G, Warner TT. CSN complex controls the stability of selected synaptic proteins via a torsinA-dependent process. EMBO J 2011; 30: 181-93. PMID: 21102408
- Hunt RA, Edris W, Chanda PK, Nieuwenhuijsen B, Young KH. Snapin interacts with the N-terminus of regulator of G protein signaling 7. Biochem Biophys Res Commun 2003; 303: 594-9. PMID: 12659861
- Ilardi JM, Mochida S, Sheng ZH. Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat Neurosci 1999; 2: 119-24. PMID: 10195194
- Krapivinsky G, Mochida S, Krapivinsky L, Cibulsky SM, Clapham DE. The TRPM7 ion channel functions in cholinergic synaptic vesicles and affects transmitter release. Neuron 2006; 52: 485-96.PMID: 17088214
- Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O'Brien EP, Tinsley CL, Blake DJ, Spritz RA, Copeland NG, Jenkins NA, Amato D, Roe BA, Starcevic M, Dell'Angelica EC, Elliott RW, Mishra V, Kingsmore SF, Paylor RE, Swank RT. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat Genet 2003; 35: 84-9. PMID: 12923531
- Lu L, Cai Q, Tian JH, Sheng ZH. Snapin associates with late endocytic compartments and interacts with late endosomal SNAREs. Biosci Rep 2009; 29: 261-9. PMID: 19335339
- Mistry AC, Mallick R, Fr?hlich O, Klein JD, Rehm A, Chen G, Sands JM. The UT-A1 urea transporter interacts with snapin, a SNARE-associated protein. J Biol Chem 2007; 282: 30097-106. PMID: 17702749
- Mistry AC, Mallick R, Klein JD, Weimbs T, Sands JM, Fr?hlich O. Syntaxin specificity of aquaporins in the inner medullary collecting duct. Am J Physiol Renal Physiol 2009; 297: F292-300. PMID: 19515809
- Mohl MC, Iismaa SE, Xiao XH, Friedrich O, Wagner S, Nikolova-Krstevski V, Wu J, Yu ZY, Feneley M, Fatkin D, Allen DG, Graham RM. Regulation of murine cardiac contractility by activation of {alpha}1A-adrenergic receptor-operated Ca2+ entry. Cardiovasc Res 2011; [Epub ahead of print] PMID: 21546445
- Morenilla-Palao C, Planells-Cases R, Garcia-Sanz N, Ferrer-Montiel A. Regulated exocytosis contributes to protein kinase C potentiation of vanilloid receptor activity. J Biol Chem 2004; 279: 25665-72. PMID: 15066994
- Moriyama K, Bonifacino JS. Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic 2002; 3: 666-77. PMID: 12191018
- Nazarian R, Starcevic M, Spencer MJ, Dell'Angelica EC. Reinvestigation of the dysbindin subunit of BLOC-1 (biogenesis of lysosome-related organelles complex-1) as a dystrobrevin-binding protein. Biochem J 2006; 395: 587-98. PMID: 16448387
- Pan PY, Tian JH, Sheng ZH. Snapin facilitates the synchronization of synaptic vesicle fusion. Neuron 2009; 61: 412-24.PMID: 19217378
- Ruder C, Reimer T, Delgado-Martinez I, Hermosilla R, Engelsberg A, Nehring R, Dorken B, Rehm A. EBAG9 adds a new layer of control on large dense-core vesicle exocytosis via interaction with Snapin. Mol Biol Cell 2005; 16: 1245-57.PMID: 15635093
- Schaaf CP, Benzing J, Schmitt T, Erz DH, Tewes M, Bartram CR, Janssen JW. Novel interaction partners of the TPR/MET tyrosine kinase. FASEB J 2005; 19: 267-9. PMID: 15546961
- Song WJ, Seshadri M, Ashraf U, Mdluli T, Mondal P, Keil M, Azevedo M, Kirschner LS, Stratakis CA, Hussain MA. Snapin mediates incretin action and augments glucose-dependent insulin secretion. Cell Metab 2011; 13: 308-19. PMID: 21356520
- Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J Biol Chem 2004; 279: 28393-401. PMID: 15102850
- Suzuki F, Morishima S, Tanaka T, Muramatsu I. Snapin, a new regulator of receptor signaling, augments alpha1A-adrenoceptor-operated calcium influx through TRPC6. J Biol Chem 2007; 282: 29563-73. PMID: 17684020
- Talbot K, Cho DS, Ong WY, Benson MA, Han LY, Kazi HA, Kamins J, Hahn CG, Blake DJ, Arnold SE. Dysbindin-1 is a synaptic and microtubular protein that binds brain snapin. Hum Mol Genet 2006; 15: 3041-54. PMID: 16980328
- Thakur P, Stevens DR, Sheng ZH, Rettig J. Effects of PKA-mediated phosphorylation of Snapin on synaptic transmission in cultured hippocampal neurons. J Neurosci 2004; 24: 6476-81.PMID: 15269257
- Tian JH, Wu ZX, Unzicker M, Lu L, Cai Q, Li C, Schirra C, Matti U, Stevens D, Deng C, Rettig J, Sheng ZH. The role of Snapin in neurosecretion: snapin knock-out mice exhibit impaired calcium-dependent exocytosis of large dense-core vesicles in chromaffin cells. J Neurosci 2005; 25: 10546-55. PMID: 16280592
- Vites O, Rhee JS, Schwarz M, Rosenmund C, Jahn R. Reinvestigation of the role of snapin in neurotransmitter release. J Biol Chem 2004; 279: 26251-6. PMID: 15084593
- Wei S, Xu Y, Shi H, Wong SH, Han W, Talbot K, Hong W, Ong WY. EHD1 is a synaptic protein that modulates exocytosis through binding to snapin. Mol Cell Neurosci 2010; 45: 418-29. PMID: 20696250
- Wolff S, Stoter M, Giamas G, Piesche M, Henne-Bruns D, Banting G, Knippschild U. Casein kinase 1 delta (CK1delta) interacts with the SNARE associated protein snapin. FEBS Lett 2006; 580: 6477-84. PMID: 17101137
- Yuan X, Shan Y, Zhao Z, Chen J, Cong Y. Interaction between Snapin and G-CSF receptor. Cytokine 2006; 33: 219-25. PMID: 16595180
- Zhou B, Zhu YB, Lin L, Cai Q, Sheng ZH. Snapin deficiency is associated with developmental defects of the central nervous system. Biosci Rep 2011; 31: 151-8. PMID: 20946101
- Zissimopoulos S, West DJ, Williams AJ, Lai FA. Ryanodine receptor interaction with the SNARE-associated protein snapin. J Cell Sci 2006; 119: 2386-97.PMID: 16723744
EDIT HISTORY:
Created by Wei Li, 07/12/2004
Updated by Wei Li, 06/25/2005
Updated by Wei Li, 12/10/2005
Updated by Wei Li, 12/28/2006
Updated by Wei Li, 02/28/2008
Updated by Wei Li, 03/12/2009
Updated by Wei Li, 05/26/2011