GENOMIC
Mapping
11p15.1. View the map and BAC clones of FISH (data from UCSC genome browser).
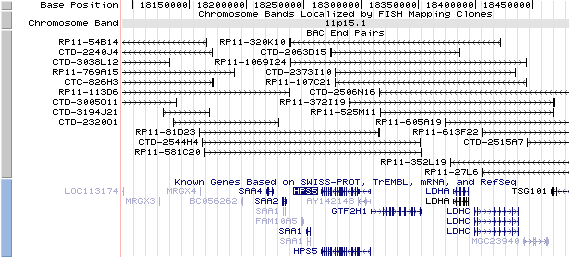
Structure
(assembly 07/03)
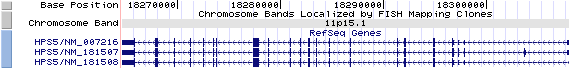
Isoform a (NM_181507): 23 exons, 43,505 bp, chr11:18,264,526-18,308,030.
Isoform b (NM_007216): 22 exons, 43,505 bp, chr11:18,264,526-18,308,030.
Isoform b (NM_181508): 21 exons, 43,505 bp, chr11:18,264,526-18,308,030.
1) NM_007216: variant lacks an in-frame segment of the coding region, compared to isoform a. These differences cause translation initiation at a downstream ATG and an isoform b with a shorter N-terminus compared to isoform a.
2) NM_181508: variant differs in the 5' UTR and lacks an in-frame segment of the coding region, compared to isoform a. These differences, similiar to those in NM_007216, can also cause translation initiation at a downstream ATG and an isoform b with a shorter N-terminus compared to isoform a.
The figure shows the comparison of the three isoforms (data from UCSC genome browser).
Regulatory Element
Search the 5'UTR upstream regions (human and mouse) of isoform (a) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
a) Transcript variant 1 (NM_181507), 4,880bp, view ORF and the alignment to genomic.
b) Transcript variant 2 (NM_007216), 4,723bp, view ORF and the alignment to genomic.
c) Transcript variant 3 (NM_181508), 4,536bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: ubiquitously expressed, with highest expression in liver, spinal chord, testis and thalamus.
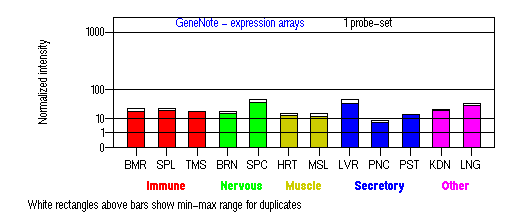
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle; LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
(HPS5p)
Isoform a: ( NP_852608): 1129aa, ExPaSy NiceProt view of Swiss-Prot:Q9UPZ3.
Isoform b ( NP_852609): 1015aa.
Isoform b ( NP_009147): identical to NP_852609.
Synonyms: Hermansky-Pudlak syndrome 5 protein; Alpha-integrin-binding protein 63;
Ruby-eye protein 2 homolog.
Ortholog
| Species | Mouse | Rat | Chicken | Fugufish | Drosophila |
| GeneView | ru2/Hps5 | LOC308598 | 06266 | Q7M564 | CG9770 |
| Protein | AA025958 (1126aa) | XP_218589 (903aa) | 10114 (1138aa) | Q7M564 (1110aa) | Q8SXI1 (826aa) |
| Identities | 81%/918aa | 73%/700aa | 66%/757aa | 49%/568aa | 29%/128aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) CDD domain: KOG3621.
(2) WD40 domains: 31-71, 72-113.
(3) Domains predicted by SMART(isoform a):
a) low complexity: 20-35
b) low complexity: 47-58
c) low complexity: 429-449
d) low complexity: 590-599
e) low complexity: 777-788
f) low complexity: 992-1001
(4) Transmembrane domains predicted by SOSUI(isoform a): none.
(5) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite(isoform a):
a) N-glycosylation site [pattern] [Warning: pattern with a high probability of occurrence]:
153 - 156 NTSK, 505 - 508 NVSH, 518 - 521 NETF, 664 - 667 NSSM, 677 - 680 NESK, 1050 - 1053 NGSL.
b) Tyrosine sulfation site [rule] [Warning: rule with a high probability of occurrence]:
82 - 96 cclhdddYvavatsq, 212 - 226 nkerdgeYgacffpg.
c) cAMP- and cGMP-dependent protein kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
124 - 127 RRvT,
434 - 437 RRsS,
458 - 461 RRgS,
551 - 554 RKtT,
692 - 695 KRdS.
d)Tyrosine kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
109 - 116 RgkpEqm.Y,
213 - 219 Ker.Dge.Y,
402 - 410 KsqlDhgtY.
e) Amidation site [pattern]:
122 - 125 kGRR.
f) Cell attachment sequence [pattern] [Warning: pattern with a high probability of occurrence]:
573 - 575 RGD.
g) Serine-rich region [profile]:
429 - 476 SacssrrssisshesfsildsgiyriissrrgsqsdedscslhsqtlS.
(2) Predicted results of subprograms by PSORT II(isoform a):
a) N-terminal signal peptide: none, N-terminal side will be inside.
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase: none.
(2) 3D models of isoform a predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
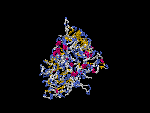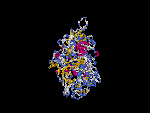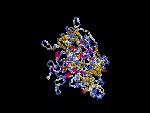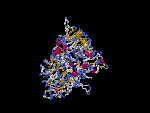
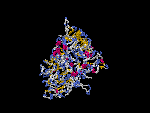
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=127,449Da, pI=5.35 (isoform a).
Computed theoretical MW=114,826Da, pI=5.24 (isoform b).
FUNCTION
Ontology
a) Biological process: visual perception.
b) Might be involved in the regulation of general functions of integrins (Wixler, et al).
c) Might be related to the trafficking of cholestrol-containing vesicles (Huizing, et al).
Location
Cytoplasmic.
Interaction
Zhang, et al reported that HPS5p directly interacts with HPS6p in a complex known as biogenesis of lysosome-related organelles complex-2 (or BLOC2). Gautum, et al found that HPS3p resides in the same protein complex. The native molecular mass of BLOC-2 was estimated to be 340 +/- 64 kDa (view diagram of BLOC-2 complex here). BLOC-2 exists in a soluble pool and associates to membranes as a peripheral membrane protein (Di Pietro, et al).
HPS5 might interact with all alpha-integrin chains that have an aromatic residue before the first lysine of the conserved KXGFFKR motif, including ITGA2, ITGA3, ITGA5 and ITGA6 (Wixler, et al).
No interactions found in the CuraGen database by searching its drosophila homolog CG9770.The drosophila pigment mutant pink (p) is identified as the homologue of HPS5 (Falcon-Perez, et al).
Pathway
May regulate the synthesis and function of lysosomes and of highly specialized organelles, such as melanosomes and platelet dense granules (view diagram of BLOC-2 pathway here).
MUTATION
Allele or SNP
No mutations have been deposited in HGMD.
SNPs deposited in dbSNP.
1 allelic variant described in OMIM.
Distribution
| Location | Genomic | cDNA | Protein | Type | Ethnicity | Reference |
| Exon 5 | 302_305delTTTG | 302_305delTTTG | V101delTTTG | frame-shift, 103X | Cuban-Venezuelan | Carmona-Rivera, et al |
| Exon 8 | 877insC | 877insC | P293insC | frame-shift | English, Irish | Huizing, et al |
| Exon 8 | 888insA | 888insA | H297insA | frame-shift | Turkish | Korswagen, et al |
| Exon 12 | 1423delC | 1423delC | L475delC | frame-shift, 511X | Mexican | Carmona-Rivera, et al |
| Intron 13 | 1634+1G>A | 1634+1G>A | 1634+1G>A | splicing, skipping E13 (124bp) | Cuban-Venezuelan | Carmona-Rivera, et al |
| Exon 16 | 1871T>G | 1871T>G | L624R | missense | Swiss | Huizing, et al |
| Exon 16 | 2024delAGTT | 2024delAGTT | L675delAGTT | frame-shift 683X | Turkish | Zhang, et al |
| Exon 18 | 2593C>T | 2593C>T | R865X | nonsense | English, Irish, Dutch, Sweden | Huizing, et al |
| Exon 18 | 2623delT | 2623delT | L875delT | frame-shift 894X | English, Irish, Dutch, Sweden | Huizing, et al |
| Exon 20 | 2929insGA | 2929insGA | T977insGA | frame-shift | English, Irish | Huizing, et al |
| Exon 22 | 3293C>T | 3293C>T | T1098I | missense | Swiss | Huizing, et al |
(Numbering of genomic and cDNA sequence is based on the start codon of RefSeq NM_181507. The P239insC mutation is not defined here because of the confused location.)
Effect
The L675delAGTT mutation in HPS5 is predicted to be a protein null mutation. Huizing, et al reported that the two HPS-5 patients with nonsense or frame shift mutations showed severely decreased HPS5 mRNA, attributable to nonsense mediated decay.
PHENOTYPE
Defects in HPS5 are the cause of Hermansky-Pudlak syndrome type 5 (HPS-5, OMIM 607521). Zhang, et al described an HPS-5 3-year-old patient with clinically mild oculocutaneous albinism and easy bruising. His platelet count was moderately reduced, and his bleeding time was moderately prolonged. Huizing, et al reported the HPS-5 patients, similar to HPS-6 patients, exhibited iris transillumination, variable hair and skin pigmentation, absent platelet dense bodies, but without pulmonary fibrosis and granulomatous colitis. The HPS5 patients also have increased serum cholestrol levels. In all the four HPS5 patients they described, LAMP-3 distribution was restricted to the perinuclear region, and was strikingly decreased in the dendritic tips of fibroblasts. Tyrosinase and TYRP1 are mistrafficked and fail to be efficiently delivered to melanosomes of HPS-5 melanocytes. However, early stage melanosome formation and Pmel17 trafficking are preserved in HPS5-deficient cells (Helip-Wooley, et al).
REFERENCE
- Carmona-Rivera C, Golas G, Hess RA, Cardillo ND, Martin EH, O'Brien K, Tsilou E, Gochuico BR, White JG, Huizing M, Gahl WA. Clinical, molecular, and cellular features of non-Puerto Rican Hermansky-Pudlak syndrome patients of Hispanic descent. J Invest Dermatol 2011; 131: 2394-400. PMID: 21833017
- Di Pietro SM, Falcon-Perez JM, Dell'Angelica EC. Characterization of BLOC-2, a complex containing the Hermansky-Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic 2004; 5: 276-83. PMID: 15030569
- Falcon-Perez JM, Romero-Calderon R, Brooks ES, Krantz DE, Dell'angelica EC. The Drosophila Pigmentation Gene pink (p) Encodes a Homologue of Human Hermansky-Pudlak Syndrome 5 (HPS5).Traffic, 2007; 8: 154-68. PMID: 17156100
- Gautam R, Chintala S, Li W, Zhang Q, Tan J, Novak EK, Di Pietro SM, Dell'Angelica EC, Swank RT. The Hermansky-Pudlak syndrome 3 (cocoa) protein is a component of the biogenesis of lysosome-related organelles complex-2 (BLOC-2). J Biol Chem 2004; 279: 12935-42. PMID: 14718540
- Helip-Wooley A, Westbroek W, Dorward HM, Koshoffer A, Huizing M, Boissy RE, Gahl WA. Improper trafficking of melanocyte-specific proteins in Hermansky-Pudlak syndrome type-5. J Invest Dermatol 2007; 127:1471-8. PMID: 17301833
- Huizing M, Hess R, Dorward H, Claassen DA, Helip-Wooley A, Kleta R, Kaiser-Kupfer MI, White JG, Gahl WA. Cellular, molecular and clinical characterization of patients with hermansky-pudlak syndrome type 5. Traffic 2004; 5: 711-22. PMID: 15296495
- Korswagen LA, Huizing M, Simsek S, Janssen JJ, Zweegman S. A novel mutation in a Turkish patient with Hermansky-Pudlak syndrome type 5. Eur J Haematol 2008; 80: 356-60. PMID: 18182080
- Wixler V, Laplantine E, Geerts D, Sonnenberg A, Petersohn D, Eckes B, Paulsson M, Aumailley M. Identification of novel interaction partners for the conserved membrane proximal region of alpha-integrin cytoplasmic domains. FEBS Lett 1999; 445: 351-5. PubMed ID : 10094488
- Zhang Q, Zhao B, Li W, Oiso N, Novak EK, Rusiniak ME, Gautam R, Chintala S, O'Brien EP, Zhang Y, Roe BA, Elliott RW, Eicher EM, Liang P, Kratz C, Legius E, Spritz RA, O'Sullivan TN, Copeland NG, Jenkins NA, Swank RT. Ru2 and Ru encode mouse orthologs of the genes mutated in human Hermansky-Pudlak syndrome types 5 and 6. Nat Genet 33: 145-54, 2003. PubMed ID : 12548288
EDIT HISTORY:
Created by Wei Li & Jonathan W. Bourne: 06/17/2004
Updated by Wei Li: 08/18/2004
Updated by Wei Li: 02/22/2008
Updated by Wei Li: 04/14/2008
Updated by Wei Li: 06/28/2012
Updated by Wei Li: 06/13/2013