GENOMIC
Mapping
5qc3.3. View the map and BAC clones (data from UCSC genome browser).
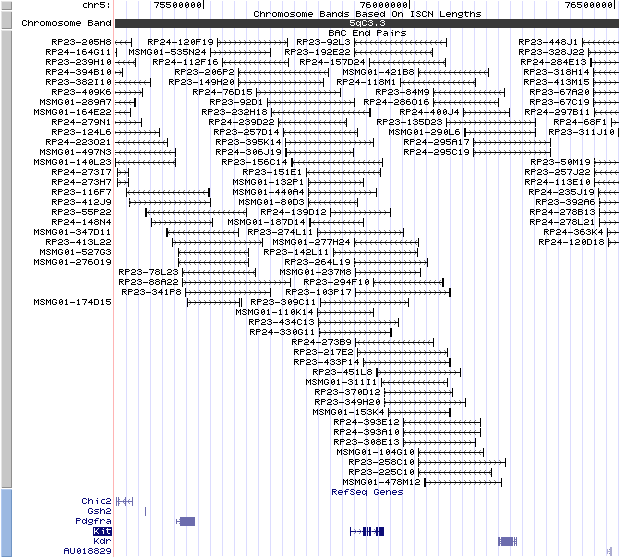
Structure
(assembly 02/2006)
Kit (NM_021099): 21 exons, 81,696 bp, chr5:75856721-75938416.
The figure below shows the structure of the Kit gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Kit, seq2=human KIT) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Kit (NM_021099), 5,132 bp, view ORF and the alignment to genomic.
Expression Pattern
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
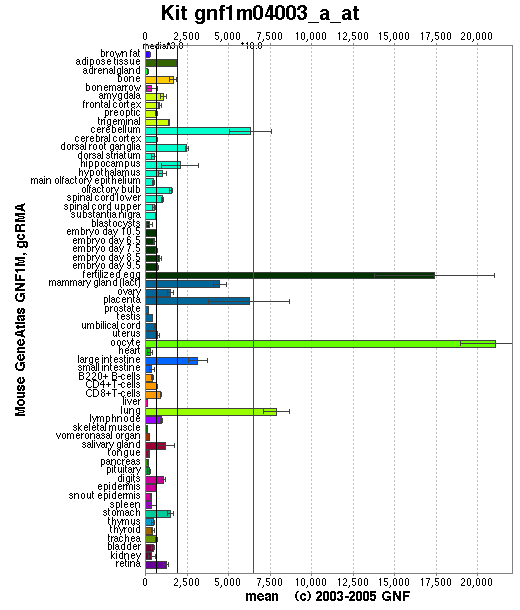
Browse more information in Entrez Gene, UCSC Gene Sorter, MGI.
PROTEIN
Sequence
c-kit (NP_066922) : 975 aa, UniProtKB/Swiss-Prot entry P05532.
This isoform has an alternate splice site in the CDS and encodes a shorter isoform, as compared to longer isoform (979aa, NP_001116205) encoded by NM_001122733, of which two are characterized by the presence or absence of four amino acids (GNNK- and GNNK+, respectively) in the extracellular domain.
Ortholog
Species
Human Chimpanzee Dog Rat Chicken GeneView
KIT
KIT
KIT
Kit
KIT
Protein
NP_000213(976 aa)
XP_517285 (1,073 aa)
NP_001003181 (975 aa)
NP_071600 (978 aa)
NP_989692 (960 aa)
Identities
800/979 (81%) 803/975 (82%) 798/977 (81%) 898/979 (91%) 613/939 (65%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) signal peptide: 1-24
b) low complexity: 25-38
c) IG_like: 49-105
d) IG_like: 122-206
e) IGc2: 225-300
f) IG_like: 323-413
g) IG_like: 429-501
h) transmembrane: 520-542
i) TyrKc: 588-922
j) low complexity: 941-959
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a MEMBRANE PROTEIN
which have 2 transmembrane helices.
No. N terminal transmembrane region C terminal type length 1 1 MRGARGAWDLLCVLLVLLRG 20 SECONDARY 20 2 520 LFTPLLIGFVVAAGAMGIIVMVL 542 PRIMARY 23
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 146 to 149 NYSL.
Site : 296 to 299 NNTF.
Site : 303 to 306 NVTT.
Site : 323 to 326 NTTV.
Site : 355 to 358 NRTS.
Site : 370 to 373 NKSN.
Site : 466 to 469 NVSV.
Site : 489 to 492 NGTV.
Site : 817 to 820 NDSN.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 684 to 687 KRDS.
Site : 736 to 739 KRRS.
Site : 911 to 914 KRPT.
c) Protein kinase C phosphorylation site:
Site : 67 to 69 TFK.
Site : 172 to 174 TIK.
Site : 198 to 200 SDK.
Site : 202 to 204 TLK.
Site : 235 to 237 TIK.
Site : 282 to 284 SAR.
Site : 307 to 309 TLK.
Site : 543 to 545 TYK.
Site : 631 to 633 TER.
Site : 734 to 736 TDK.
Site : 739 to 741 SAR.
Site : 914 to 916 TFK.
Site : 929 to 931 STK.
Site : 953 to 955 SVR.
d) Casein kinase II phosphorylation site:
Site : 30 to 33 SPGE.
Site : 57 to 60 TCID.
Site : 148 to 151 SLIE.
Site : 216 to 219 SVPE.
Site : 235 to 238 TIKD.
Site : 329 to 332 TDGE.
Site : 398 to 401 SNSD.
Site : 414 to 417 TKPE.
Site : 447 to 450 TGAE.
Site : 458 to 461 SPVD.
Site : 479 to 482 SSID.
Site : 631 to 634 TERE.
Site : 691 to 694 SKQE.
Site : 715 to 718 SSNE.
Site : 732 to 735 TKTD.
Site : 744 to 747 SYIE.
Site : 903 to 906 TCWD.
e) Tyrosine kinase phosphorylation site:
Site : 386 to 393 KGTEGGTY.
Site : 601 to 608 KVVEATAY.
f) N-myristoylation site:
Site : 3 to 8 GARGAW.
Site : 20 to 25 GQTATS.
Site : 94 to 99 GTYTCS.
Site : 103 to 108 GLTSSI.
Site : 289 to 294 GVFMCY.
Site : 387 to 392 GTEGGT.
Site : 390 to 395 GGTYTF.
Site : 427 to 432 GMLQCV.
Site : 490 to 495 GTVECK.
Site : 609 to 614 GLIKSD.
Site : 647 to 652 GNHMNI.
Site : 960 to 965 GSSASS.
g) Cell attachment sequence:
Site : 267 to 269 RGD.
h) ATP/GTP-binding site motif A (P-loop):
Site : 496 to 503 ASNDVGKS.
i) Protein kinases ATP-binding region signature:
Site : 594 to 622 LGAGAFGKVVEATAYGLIKSDAAMTVAVK.
j) Tyrosine protein kinases specific active-site signature:
Site : 786 to 798 CIHRDLAARNILL.
k) Receptor tyrosine kinase class III signature:
Site : 647 to 660 GNHMNIVNLLGACT.
(2) Predicted results of subprograms by PSORT II:
a) Seems to have a cleavable signal peptide (1 to 22)
b) Tentative number of TMS(s) for the threshold 0.5: 2
Number of TMS(s) for threshold 0.5: 1
INTEGRAL Likelihood = -9.55 Transmembrane 525 - 541
PERIPHERAL Likelihood = 2.33 (at 110)
ALOM score: -9.55 (number of TMSs: 1)
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: XXRR-like motif in the N-terminus: RGAR
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: too long tail
j) Dileucine motif in the tail: found
LL at 655
LL at 677
LL at 705
LL at 767
LL at 797
LL at 969
3D Model
ModBase predicted 3D structure of P05532 from UCSC Gene Sorter:

From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=109,001.4Da, pI=6.60.
| Species | Human | Chimpanzee | Dog | Rat | Chicken |
| GeneView | KIT | KIT | KIT | Kit | KIT |
| Protein | NP_000213(976 aa) | XP_517285 (1,073 aa) | NP_001003181 (975 aa) | NP_071600 (978 aa) | NP_989692 (960 aa) |
| Identities | 800/979 (81%) | 803/975 (82%) | 798/977 (81%) | 898/979 (91%) | 613/939 (65%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) signal peptide: 1-24
b) low complexity: 25-38
c) IG_like: 49-105
d) IG_like: 122-206
e) IGc2: 225-300
f) IG_like: 323-413
g) IG_like: 429-501
h) transmembrane: 520-542
i) TyrKc: 588-922
j) low complexity: 941-959
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a MEMBRANE PROTEIN
which have 2 transmembrane helices.
| No. | N terminal | transmembrane region | C terminal | type | length |
| 1 | 1 | MRGARGAWDLLCVLLVLLRG | 20 | SECONDARY | 20 |
| 2 | 520 | LFTPLLIGFVVAAGAMGIIVMVL | 542 | PRIMARY | 23 |
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 146 to 149 NYSL.
Site : 296 to 299 NNTF.
Site : 303 to 306 NVTT.
Site : 323 to 326 NTTV.
Site : 355 to 358 NRTS.
Site : 370 to 373 NKSN.
Site : 466 to 469 NVSV.
Site : 489 to 492 NGTV.
Site : 817 to 820 NDSN.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 684 to 687 KRDS.
Site : 736 to 739 KRRS.
Site : 911 to 914 KRPT.
c) Protein kinase C phosphorylation site:
Site : 67 to 69 TFK.
Site : 172 to 174 TIK.
Site : 198 to 200 SDK.
Site : 202 to 204 TLK.
Site : 235 to 237 TIK.
Site : 282 to 284 SAR.
Site : 307 to 309 TLK.
Site : 543 to 545 TYK.
Site : 631 to 633 TER.
Site : 734 to 736 TDK.
Site : 739 to 741 SAR.
Site : 914 to 916 TFK.
Site : 929 to 931 STK.
Site : 953 to 955 SVR.
d) Casein kinase II phosphorylation site:
Site : 30 to 33 SPGE.
Site : 57 to 60 TCID.
Site : 148 to 151 SLIE.
Site : 216 to 219 SVPE.
Site : 235 to 238 TIKD.
Site : 329 to 332 TDGE.
Site : 398 to 401 SNSD.
Site : 414 to 417 TKPE.
Site : 447 to 450 TGAE.
Site : 458 to 461 SPVD.
Site : 479 to 482 SSID.
Site : 631 to 634 TERE.
Site : 691 to 694 SKQE.
Site : 715 to 718 SSNE.
Site : 732 to 735 TKTD.
Site : 744 to 747 SYIE.
Site : 903 to 906 TCWD.
e) Tyrosine kinase phosphorylation site:
Site : 386 to 393 KGTEGGTY.
Site : 601 to 608 KVVEATAY.
f) N-myristoylation site:
Site : 3 to 8 GARGAW.
Site : 20 to 25 GQTATS.
Site : 94 to 99 GTYTCS.
Site : 103 to 108 GLTSSI.
Site : 289 to 294 GVFMCY.
Site : 387 to 392 GTEGGT.
Site : 390 to 395 GGTYTF.
Site : 427 to 432 GMLQCV.
Site : 490 to 495 GTVECK.
Site : 609 to 614 GLIKSD.
Site : 647 to 652 GNHMNI.
Site : 960 to 965 GSSASS.
g) Cell attachment sequence:
Site : 267 to 269 RGD.
h) ATP/GTP-binding site motif A (P-loop):
Site : 496 to 503 ASNDVGKS.
i) Protein kinases ATP-binding region signature:
Site : 594 to 622 LGAGAFGKVVEATAYGLIKSDAAMTVAVK.
j) Tyrosine protein kinases specific active-site signature:
Site : 786 to 798 CIHRDLAARNILL.
k) Receptor tyrosine kinase class III signature:
Site : 647 to 660 GNHMNIVNLLGACT.
(2) Predicted results of subprograms by PSORT II:
a) Seems to have a cleavable signal peptide (1 to 22)
b) Tentative number of TMS(s) for the threshold 0.5: 2
Number of TMS(s) for threshold 0.5: 1
INTEGRAL Likelihood = -9.55 Transmembrane 525 - 541
PERIPHERAL Likelihood = 2.33 (at 110)
ALOM score: -9.55 (number of TMSs: 1)
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: XXRR-like motif in the N-terminus: RGAR
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: too long tail
j) Dileucine motif in the tail: found
LL at 655
LL at 677
LL at 705
LL at 767
LL at 797
LL at 969
3D Model
ModBase predicted 3D structure of P05532 from UCSC Gene Sorter:



2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=109,001.4Da, pI=6.60.
FUNCTION
Ontology
(1) Biological process: hemopoiesis, epithelial cell proliferation, neural crest cell migration, signal transduction, spermatogenesis, protein kinase cascade.
(2) Cytokine-mediated signaling pathway.
(3) Receptor activity.
(4) Receptor signaling protein tyrosine kinase activity, ATP binding.
(5) Regulation of pigmentation during development.
Location
Plasma membrane, cytoplasm, nucleus.
Interaction
C-kit was first identified as the cellular homolog of the feline sarcoma viral oncogene v-kit. This protein is a type 3 transmembrane receptor for MGF (mast cell growth factor, also known as stem cell factor). It has a tyrosine-protein kinase activity. Binding of the ligands leads to the autophosphorylation of KIT and its association with substrates such as phosphatidylinositol 3-kinase (Pi3K).
View interactions in HPRD
View co-occured partners in literature searched by PPI Finder.
Pathway
Mast/stem cell growth factor receptor precursor (EC 2.7.10.1) (SCFR) (Proto-oncogene tyrosine-protein kinase Kit) (c-kit) (CD117 antigen).
Cytokine-cytokine receptor interaction , Melanogenesis, Hematopoietic cell lineage in KEGG.
MUTATION
Allele or SNP
97 phenotypic alleles of Kit are described in MGI:96677.
SNPs deposited in dbSNP Build 128.
Distribution
Among the reported 97 phenotypic alleles, 66 are spontaneous and 17 are chemically induced.
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_001122733. view ORF here.)
Effect
The two isoforms show differences in signal transduction and biological activities and the shorter isoform seems to be highly expressed than the longer isoform in human malignancies. Substitution of aspartic acid with valine (D816V) renders the receptor independent of ligand for activation and signaling. The differences in downstream signal transduction and biological responses between the two GNNK isoforms are eliminated by the D816V mutant (Pedersen et al.).
PHENOTYPE
Mutations at this locus affect migration of embryonic stem cell populations, resulting in mild to severe impairments in hemopoiesis, and pigmentation. Some alleles are homozygous lethal, sterile, or result in the formation of gastrointestinal tumors. The W-v allele arose spontaneously in the C57BL/6J strain. The strain is described in more detail in JAX Mice database (C57BL/6J-KitW-v/J).
REFERENCE
- Pedersen M, Ronnstrand L, Sun J. The c-Kit/D816V mutation eliminates the differences in signal transduction and biological responses between two isoforms of c-Kit. Cell Signal 2009; 21:413-8. PMID: 19049823