GENOMIC
Mapping
7qB3. View the map and BAC cotig (data from UCSC genome browser).
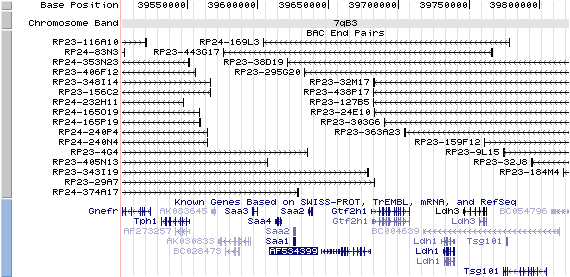
Structure
(assembly 10/03)
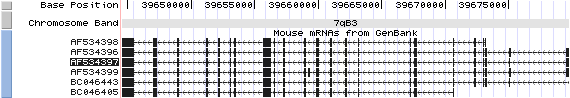
Isoform a (AF534396): 23 exons, 4,625 bp, chr7:39644540-39679750.
Isoform a (AF534397): 23 exons, 4,601 bp, chr7:39644540-39679750.
Isoform b (AF534398): 21 exons, 4,190 bp, chr7:39644540-39673243.
Isoform c (AF534399): 24 exons, 4,756 bp, chr7:39644540-39679750.
Alternative splicing in kidney(Zhang, et al):
1) AF534397: alternatively spliced; contains shorter exon 2 than AF534396, encodes same isoform (a) because it does not alter the ORF.
2) AF534398: alternatively spliced; lacks of exon 1 and contains partial exon 2 compared with AF534396, utilizes alternative ATG start site to encode isoform (b) (truncated by 165aa at N-terminal isoform a).
3) AF534399: alternatively spliced; contains additional exon between exons 12 and 13 compared with AF534396, encoding isoform (c) (truncated by 600aa at C-terminal isoform a).
The figure shows the comparison of these 4 variants (data from UCSC genome browser).
Regulatory Element
Search the 5'UTR regions (human and mouse) of isoform (a) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
a) Transcript variant 1 (AF534396), 4,625bp, view ORF and the alignment to genomic.
b) Transcript variant 2 (AF534397), 4,601bp, view ORF and the alignment to genomic.
c) Transcript variant 3 (AF534398), 4,190bp, view ORF and the alignment to genomic.
d) Transcript variant 4 (AF534399), 4,756bp, view ORF and the alignment to genomic.
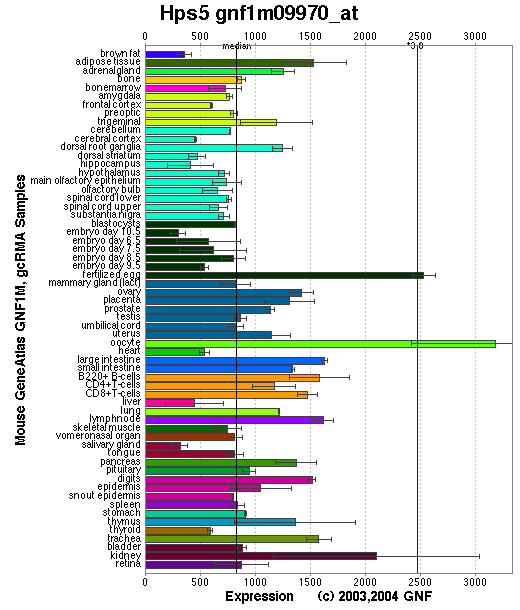
Expression Pattern
Tissue specificity: widely expressed, with lowest expression in skeletal muscle and spleen (Zhang, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
(Ru2/Hps5p)
Isoform a (AAO25958), 1126 aa, ExPaSy NiceProt view of Swiss-Prot: P59438.
Isoform b (AAO25960), 961 aa, ExPaSy NiceProt view of Swiss-Prot: P59438.2.
Isoform c (AAO25961), 526 aa, ExPaSy NiceProt view of Swiss-Prot: P59438.3.
Synonyms: Ruby-eye protein 2; Hermansky-Pudlak syndrome 5 protein homolog.
Ortholog
| Species | Human | Rat | Chicken | Fugufish | Drosophila |
| GeneView | HPS5 | LOC308598 | 06266 | Q7M564 | CG9770 |
| Protein | NP_852608 (1129aa) | XP_218589 (903aa) | 10114 (1138aa) | Q7M564 (1110aa) | Q8SXI1 (826aa) |
| Identities | 81%/918aa | 86%/811aa | 64%/735aa | 49%/570aa | 32%/105aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART(isoform a):
a) low complexity: 429-449
b) low complexity: 775-786
c) low complexity: 989-998
d) low complexity: 1021-1033
(2) Transmembrane domains predicted by SOSUI(isoform a): none.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite(isoform a):
a) N-glycosylation site [pattern] [Warning: pattern with a high probability of occurrence]:
659 - 662 NTSS, 710 - 713 NLSE, 1047 - 1050 NGSL.
b) Tyrosine sulfation site [rule] [Warning: rule with a high probability of occurrence]:
82 - 96 ccshdddYvavatsq, 812 - 826 marsnpaYteleegd.
c) Glycosaminoglycan attachment site [rule] [Warning: rule with a high probability of occurrence]:
686 - 689 SGeG.
d) cAMP- and cGMP-dependent protein kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
124 - 127 RKvT,
434 - 437 RRsS,
458 - 461 RRgS,
551 - 554 RKtT,
690 - 693 RRvS.
e)Tyrosine kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
328 - 336 KdiqDvavY.
e) Amidation site [pattern]:
122 - 125 kGRK, 688 - 691 eGRR.
f) Serine-rich region [profile]:
429 - 476 SacssrrssisshvsfsildsgiyriissrrgsqsdedscslhsqtfS.
(2) Predicted results of subprograms by PSORT II(isoform a):
a) N-terminal signal peptide: none, N-terminal side will be inside.
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase: none.
(2) 3D models of isoform (a) predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.

2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=126,307Da, pI=5.54 (isoform a).
Computed theoretical MW=108,189Da, pI=5.22 (isoform b).
Computed theoretical MW= 59,028Da, pI=5.54 (isoform c).
FUNCTION
Ontology
(1) Biological process: visual perception
(2) May regulate the synthesis and function of lysosomes and of highly specialized organelles, such as melanosomes and platelet dense granules (view diagram of BLOC-2 pathway here).
Location
Cytoplasmic.
Interaction
Zhang, et al reported that HPS5p directly interacts with HPS6p in a complex known as biogenesis of lysosome-related organelles complex-2 (or BLOC2). Gautum, et al found that HPS3p resides in the same protein complex. The native molecular mass of BLOC-2 was estimated to be 340 +/- 64 kDa (view diagram of BLOC-2 complex here). BLOC-2 exists in a soluble pool and associates to membranes as a peripheral membrane protein (Di Pietro, et al).
No interactions found in the CuraGen database by searching its drosophila homolog CG9770.
Pathway
During the maturation of melanosomes, The BLOC-2 mutants ru2 and ru are blocked between stages I and II ( Nguyen, et al ) (view diagram of melanosome blockage here).
MUTATION
Allele or SNP
6 phenotypic alleles described in MGI:2180307.
SNPs deposited in dbSNP.
Distribution
| Location | Genomic | cDNA | Protein | Type | Strain | Reference |
| Exon 10 | 997delG | 997delG | V333delG | frame-shift 333X | ru2d (B6) | Hirobe, et al |
| Exon 16 | 2270GdelGinsTT | 2270GdelGinsTT | G757delGinsTT | frame-shift 816X | ru2J (B6) | Zhang, et al |
| Exon 18 | 2698GinsCCGG | 2698GinsCCGG | E900insCCGG | frame-shift 909X | ru2hz (DBA/2J) | Zhang, et al |
| Exon 18 | 2601Gins 1kb H2A histone, 2594-2601dup | 2601Gins 1kb H2A histone, 2594-2601dup | K867ins 1kb H2A histone | gross insertion | ru2 (B6) | Zhang, et al |
(Numbering of genomic and cDNA sequence is based on the start codon of RefSeq AF534396.)
Effect
No differences between the northern blot pattern of tissues of homozygous ru2J and those of wild type mice were observed (Zhang, et al ). The Hps5 protein is missing in Western blots in spleen and lung. The Hps5 protein also exhibits destabilization in Hps3 or Hps6 null mutants (Gautum, et al ). The ru2(d) allele inhibits melanocyte differentiation, and that its impaired differentiation is rescued by excess Tyr (Hirobe, et al ).
PHENOTYPE
Defects in Hps5 are the cause of ruby-eye 2, a mouse model of Hermansky-Pudlak syndrome type 5 (HPS-5, OMIM 607521). Homozygotes have hypopigmented eyes and hair, impaired secretion of lysosomal enzymes by renal proximal tubules and reduced clotting due to a platelet dense granule defect (Novak, et al) (more details in Mouse Locus Catalog #Hps5). The strain is described in more detail in JAX Mice database (B6-Hps5ru2-J/J). Melanosomes of the retinal pigment epithelium (RPE) of the BLOC-2 mutants Hps5/ru and Hps6/ru2 are similar, which are greatly reduced in number and those remaining appear to be degradative forms. Melanosomes of the choroid are smaller, and some are found grouped within distinctive membrane-limited organelles (Zhang, et al). The ru2 and ru mice are completely indistinguishable in appearance. In addition, Hps5/ru2-Hps6/ru doubly homozygous mouse presents a phenotype identical to that of the singly homozygous mutants (Gautum, et al).
The drosophila pigment mutant pink (p) is identified as the homologue of HPS5 (Falcon-Perez, et al).
REFERENCE
- Di Pietro SM, Falcon-Perez JM, Dell'Angelica EC. Characterization of BLOC-2, a complex containing the Hermansky-Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic 2004; 5: 276-83. PMID: 15030569
- Falcon-Perez JM, Romero-Calderon R, Brooks ES, Krantz DE, Dell'angelica EC. The Drosophila Pigmentation Gene pink (p) Encodes a Homologue of Human Hermansky-Pudlak Syndrome 5 (HPS5).Traffic, 2006 Dec 7; [Epub ahead of print]PMID: 17156100
- Gautam R, Chintala S, Li W, Zhang Q, Tan J, Novak EK, Di Pietro SM, Dell'Angelica EC, Swank RT. The Hermansky-Pudlak syndrome 3 (cocoa) protein is a component of the biogenesis of lysosome-related organelles complex-2 (BLOC-2). J Biol Chem 2004; 279: 12935-42. PMID: 14718540
- Hirobe T, Yoshihara C, Takeuchi S, Wakamatsu K, Ito S, Abe H, Kawa Y, Soma Y. A novel deletion mutation of mouse ruby-eye 2 named ru2(d)/Hps5(ru2-d) inhibits melanocyte differentiation and its impaired differentiation is rescued by L-tyrosine. Zoolog Sci 2011; 28: 790-801. PMID: 22035301
- Nguyen T, Novak EK, Kermani M, Fluhr J, Peters LL, Swank RT, Wei ML. Melanosome morphologies in murine models of hermansky-pudlak syndrome reflect blocks in organelle development. J Invest Dermatol 2002; 119: 1156-64. PMID: 12445206
- Novak EK, Wieland F, Jahreis GP, Swank RT. Altered secretion of kidney lysosomal enzymes in the mouse pigment mutants ruby-eye, ruby-eye-2-J, and maroon. Biochem Genet 1980; 18: 549-61. PMID: 6776948
- Zhang Q, Zhao B, Li W, Oiso N, Novak EK, Rusiniak ME, Gautam R, Chintala S, O'Brien EP, Zhang Y, Roe BA, Elliott RW, Eicher EM, Liang P, Kratz C, Legius E, Spritz RA, O'Sullivan TN, Copeland NG, Jenkins NA, Swank RT. Ru2 and Ru encode mouse orthologs of the genes mutated in human Hermansky-Pudlak syndrome types 5 and 6. Nat Genet 33: 145-54, 2003. PubMed ID : 12548288
EDIT HISTORY:
Created by Wei Li & Jonathan W. Bourne: 06/17/2004
Updated by Wei Li: 12/25/2006
Updated by Wei Li: 06/28/2012