GENOMIC
Mapping
3qA2. View the map and BAC contig (data from UCSC genome browser).
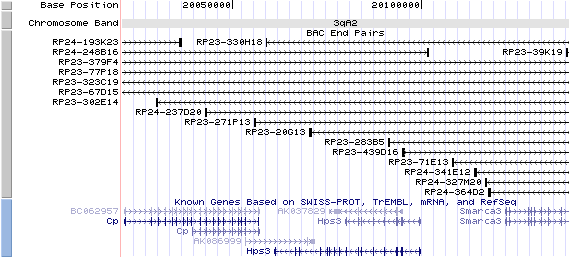
Structure
(assembly 10/03)
Hps3/NM_080634: 17 exons, 38,681 bp, chr3:20,060,835-20,099,515.
The figure shows the structure of the Hps3 gene. (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=human HPS3, seq2=mouse Hps3) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Hps3/NM_080634, 3,976 bp, view ORF and the alignment to genomic.
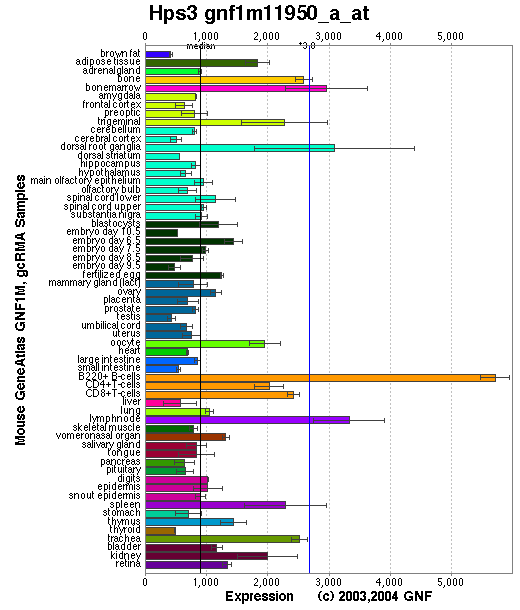
Expression Pattern
Tissue specificity: Ubiquitous.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
Hermansky-Pudlak syndrome 3 protein homolog (NP_542365):
1,002aa, ExPaSy NiceProt view of Swiss-Prot: Q91VB4.
Synonym: Cocoa protein.
Ortholog
| Species | Human | Rat | Zebrafish | Drosophila |
| GeneView | HPS3 | LOC310288 | ENSDARG00000015749 | CG14562 |
| Protein | NP_115759 (1,004aa) | XP_227003 (721aa) | ENSDARP00000018977 (1,001aa) | Q8MZ59 (1,190aa) |
| Identities | 80%/812aa | 72%/599aa | 45%/465aa | 19%/47aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) low complexity: 537-551
b) ZnF NFX: 553-563
(2) Transmembrane domains predicted by SOSUI: none (predicted as a soluble protein).
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site [pattern] [Warning: pattern with a high probability of occurrence]:
83 - 86 NKTI, 240 - 243 NETS, 490 - 493 NTTA, 609 - 612 NHSL, 852 - 855 NYTD.
b)Tyrosine sulfation site [rule] [Warning: rule with a high probability of occurrence]:
68 - 82 ayseagdYlvaieek, 400 - 414 vaheedlYmdttlka, 606 - 620 fyinhslYenldeel, 846 - 860 dsladknYtddllkl.
c) cAMP- and cGMP-dependent protein kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
457 - 460 RReS, 997 - 1000 RKkS.
d) N-myristoylation site [pattern] [Warning: pattern with a high probability of occurrence]:
226 - 231 GSmsSM, 259 - 264 GgeeNT, 345 - 350 GgedSD
e) Tyrosine kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
295 - 303 RfapDissY, 684 - 691 Kmg.DldmY
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals:
XXRR-like motif in the N-terminus: VRLY
KKXX-like motif in the C-terminus: KKSL
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: too long tail
i) Dileucine motif in the tail: found LL at 148.
3D Model
(1) ModBase: none.
(2) 3D models of isoform (a) predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
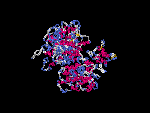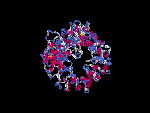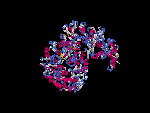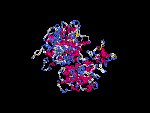
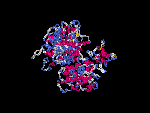
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=113,181Da, pI=5.45.
FUNCTION
Ontology
a) Biological process: organelle organization and biogenesis.
b) Component of cytoplasm.
Location
Cytoplasmic.
Interaction
Gautum, et al reported that Hps3, Hps5, and Hps6 proteins regulate vesicle trafficking to lysosome-related organelles at the physiological level as components of the BLOC-2 (biogenesis of lysosome-related organelles complex-2) protein complex. The native molecular mass of BLOC-2 was estimated to be 340 +/- 64 kDa (view diagram of BLOC-2 complex here). BLOC-2 exists in a soluble pool and associates to membranes as a peripheral membrane protein (Di Pietro, et al).
No interactions found in the CuraGen database by searching its drosophila homolog CG14562.
Pathway
Involved in early stages of melanosome biogenesis and maturation (Suzuki, et al ) ( Nguyen, et al ) (view diagram of melanosome blockage here).
MUTATION
Allele or SNP
4 phenotypic alleles described in MGI:2153839.
SNPs deposited in dbSNP.
Distribution
| Location | Genomic | cDNA | Protein | Type | Strain | Reference |
| Exon 8 | 1420-1441del 22bp | 1420-1441del 22bp | S474del 22bp | frame-shift 482X | coa3J (B10) | Suzuki, et al |
| Exon 10 | 2026ins ~5.3kb IAP, 2020-2026dup | 2026ins ~5.3kb IAP, 2020-2026dup | L676ins ~5.3kb IAP | gross insertion | coa6J (C3H/HeJ) | Suzuki, et al |
| Exon 11 | 1879A>T | 1879A>T | K627X | nonsense | coa5J (B10) | Suzuki, et al |
| Intron 14 | splicing acceptor -2A>T | 2584-2597del 14bp | S862-S866 del 14bp | frame-shift 898X | coa (B10) | Suzuki, et al |
(Numbering of genomic and cDNA sequence is based on the start codon of RefSeq NM_080634.)
Effect
All the mutations are predicted as pathological. The Hps3 transcript in coa/coa is almost invisible, although a ~9.5kb transcript was detected in the Northern blot in coa6J (Suzuki, et al).
All the mutations are predicted as pathological. The Hps3 transcript in coa/coa is almost invisible, although a ~9.5kb transcript was detected in the Northern blot in coa6J (Suzuki, et al).
PHENOTYPE
Defects in Hps3 are the cause of the cocoa (coa) mutant (Novak, et al), which is characterized by hypopigmentation and platelet dysfunction. The coa mutation arose spontaneously in the C57BL/10J strain. The strain is described in more detail in JAX Mice database (B6.B10-Hps3coa/J). The coa coat color phenotype is similar to that seen with ruby (Hps6), and coa/coa mice cannot be distinguished from either ru/ru or coa/coa-ru/ru doubly homozygous mutant mice (Gautum, et al). The mutation also causes decreased life span, increased bleeding time accompanied by symptoms of platelet storage pool deficiency (SPD), including decreased platelet serotonin and decreased visibility of dense granules as analyzed by electron microscopy of unfixed platelets. Unlike many other mouse pigment mutations that cause SPD, cocoa does not affect secretion of lysosomal enzymes. Interestingly, ApoE(-/-), HPS3(-/-) doubly mutant mice were shown to be resistant to thrombotic arterial occlusion. This suggests that the reduced platelet dense-granule secretion in coa is associated with marked protection against the development of arterial thrombosis, inflammation, and neointimal hyperplasia after vascular injury(King, et al).
REFERENCE
- Di Pietro SM, Falcon-Perez JM, Dell'Angelica EC. Characterization of BLOC-2, a complex containing the Hermansky-Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic 2004; 5: 276-83. PMID: 15030569
- Gautam R, Chintala S, Li W, Zhang Q, Tan J, Novak EK, Di Pietro SM, Dell'Angelica EC, Swank RT. The Hermansky-Pudlak syndrome 3 (cocoa) protein is a component of the biogenesis of lysosome-related organelles complex-2 (BLOC-2). J Biol Chem 2004; 279: 12935-42. PMID: 14718540
- King SM, McNamee RA, Houng AK, Patel R, Brands M, Reed GL. Platelet dense-granule secretion plays a critical role in thrombosis and subsequent vascular remodeling in atherosclerotic mice. Circulation 2009; 120: 785-91. PMID: 19687360
- Novak EK, Sweet HO, Prochazka M, Parentis M, Soble R, Reddington M, Cairo A, Swank RT. Cocoa: a new mouse model for platelet storage pool deficiency. Br J Haematol 1988; 69: 371-8. PMID: 3408670
- Suzuki T, Li W, Zhang Q, Novak EK, Sviderskaya EV, Wilson A, Bennett DC, Roe BA, Swank RT, Spritz RA. The gene mutated in cocoa mice, carrying a defect of organelle biogenesis, is a homologue of the human Hermansky-Pudlak syndrome-3 gene. Genomics 2001; 78: 30-7. PMID: 11707070
EDIT HISTORY:
Created by Wei Li: 06/17/2004
Updated by Wei Li: 07/31/2012