GENOMIC
Mapping
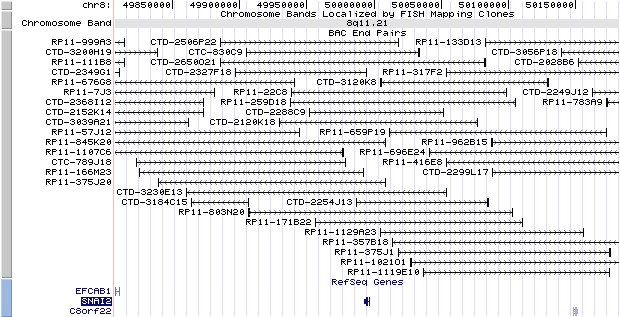
8q11.21. View the map and BAC clones (data from UCSC genome browser).
Structure
(assembly 03/2006)
SNAI2/NM_003068: 3 exons, 3,746 bp, chr8:49992796-49996541 .
The figure below shows the structure of the SNAI2 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Snai2, seq2=human SNAI2) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
SNAI2 ( NM_003068), 2,101 bp, view ORF and the alignment to genomic.
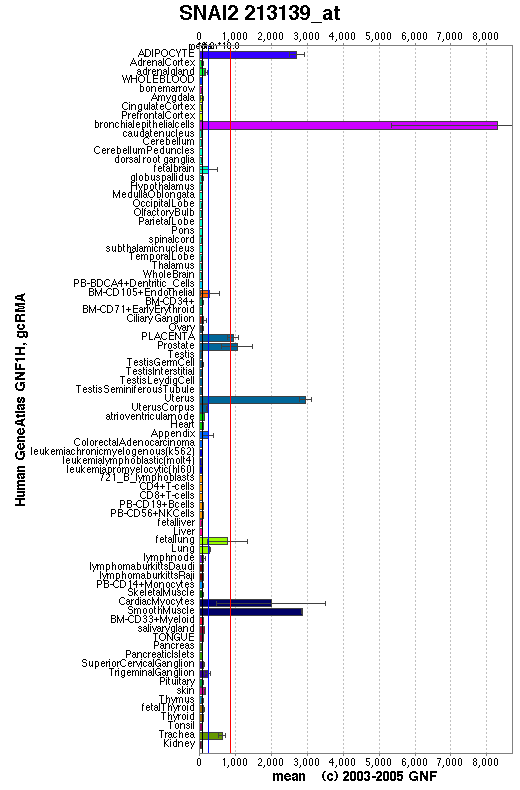
Expression Pattern
Expressed in placenta and adult heart, pancreas, liver, kidney and skeletal muscle.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
View more information about SNAI2 in Entrez Gene, or UCSC Gene Sorter, or GeneCards.
PROTEIN
Sequence
SNAI2 (snail 2) (NP_003059): 268 aa, UniProtKB/Swiss-Prot entry O43623.
Ortholog
Species
Chimpanzee Dog Mouse Fowl GeneView
SNAI2
LOC486940
Snai2
LOC421112
Protein
XP_519753.2 (268 aa)
XP_544069.1 (268 aa)
NP_035545.1 (269 aa)
XP_419196.1 (268 aa)
Identities
268/268 (100%) 262/268 (97%) 256/269 (95%) 251/268 (93%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) low complexity: 72-84
b) low complexity: 87-96
c) ZnF_C2H2: 128-150
d) ZnF_C2H2: 159-181
e) ZnF_C2H2: 185-207
f) ZnF_C2H2: 213-235
g) ZnF_C2H2: 241-268
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site:
Site : 12 to 15 NASK.
Site : 18 to 21 NYSE.
Site : 134 to 137 NKTY.
Site : 245 to 248 NCSK.
b) Protein kinase C phosphorylation site:
Site : 14 to 16 SKK.
Site : 95 to 97 SSK.
Site : 155 to 157 SRK.
Site : 160 to 162 SCK.
c) Casein kinase II phosphorylation site:
Site : 20 to 23 SELD.
Site : 95 to 98 SSKD.
Site : 100 to 103 SGSE.
Site : 107 to 110 SDEE.
Site : 234 to 237 THSD.
d) Tyrosine kinase phosphorylation site:
Site : 162 to 168 KYCDKEY.
e) N-myristoylation site:
Site : 263 to 268 GCCVAH.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of O43623 from UCSC Genome Sorter:
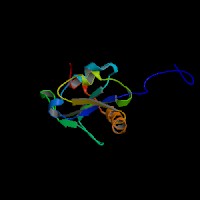
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=29, 986.1Da, pI=9.01 (NP_003059).
| Species | Chimpanzee | Dog | Mouse | Fowl |
| GeneView | SNAI2 | LOC486940 | Snai2 | LOC421112 |
| Protein | XP_519753.2 (268 aa) | XP_544069.1 (268 aa) | NP_035545.1 (269 aa) | XP_419196.1 (268 aa) |
| Identities | 268/268 (100%) | 262/268 (97%) | 256/269 (95%) | 251/268 (93%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) low complexity: 72-84
b) low complexity: 87-96
c) ZnF_C2H2: 128-150
d) ZnF_C2H2: 159-181
e) ZnF_C2H2: 185-207
f) ZnF_C2H2: 213-235
g) ZnF_C2H2: 241-268
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site:
Site : 12 to 15 NASK.
Site : 18 to 21 NYSE.
Site : 134 to 137 NKTY.
Site : 245 to 248 NCSK.
b) Protein kinase C phosphorylation site:
Site : 14 to 16 SKK.
Site : 95 to 97 SSK.
Site : 155 to 157 SRK.
Site : 160 to 162 SCK.
c) Casein kinase II phosphorylation site:
Site : 20 to 23 SELD.
Site : 95 to 98 SSKD.
Site : 100 to 103 SGSE.
Site : 107 to 110 SDEE.
Site : 234 to 237 THSD.
d) Tyrosine kinase phosphorylation site:
Site : 162 to 168 KYCDKEY.
e) N-myristoylation site:
Site : 263 to 268 GCCVAH.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of O43623 from UCSC Genome Sorter:



From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=29, 986.1Da, pI=9.01 (NP_003059).
FUNCTION
Ontology
(1) Biological process: DNA binding, DNA-dependent regulation of transcription, sensory perception of sound.
(2) Transcriptional repressor.
(3) Zinc ion binding.
(4) Involved in the generation and migration of neural crest cells. Ectoderm and mesoderm interaction.
Location
Nucleus.
Interaction
This gene encodes a member of the Snail family of C2H2-type zinc finger transcription factors. The encoded protein acts as a transcriptional repressor that binds to E-box motifs and is also likely to repress E-cadherin transcription in breast carcinoma. This protein is involved in epithelial-mesenchymal transitions and has antiapoptotic activity.
View co-occured partners in literature searched by PPI Finder.
Pathway
Adhesion Pathway in KEGG.
MUTATION
Allele or SNP
2 mutations deposited in HGMD.
SNPs deposited in dbSNP.
Selected allelic examples described in OMIM.
Distribution
Except the mutations deposited in HGMD, Sanchez-Martin, et al (2003) screened the SLUG gene mutation in 17 unrelated patients with piebaldism, that lack apparent KIT mutations. Three of these patients had evident heterozygous deletions of the SLUG gene encompassing the entire coding region.
Note that the numbering of the mutations in HGMD is based on SNAI2 cDNA (NM_003068), view ORF here.
Effect
PHENOTYPE
Defects in SNAI2 are the cause of Waardenburg syndrome type 2D (WS2D) [OMIM:608890]. WS2 is a genetically heterogeneous, autosomal dominant disorder characterized by sensorineural deafness, pigmentary disturbances, and absence of dystopia canthorum.
Some cases of piebaldism are caused by mutation in the gene encoding the zinc finger transcription factor SNAI2 [OMIM:602150]. Defects in SNAI2 are a cause of neural tube defects (NTD).
SNAI2 plays a role in the progression of some tumor types, including breast and gastric cancer. SNAI2 down-regulation facilitates apoptosis induced by proapoptotic drugs in neuroblastoma cells and decreases their invasion capability in vitro and in vivo (Vitali, et al).
REFERENCE
- Sanchez-Martin M, Perez-Losada J, Rodriguez-Garcia A, Gonzalez-Sanchez B, Korf BR, Kuster W, Moss C, Spritz RA, Sanchez-Garcia I. Deletion of the SLUG (SNAI2) gene results in human piebaldism. Am J Med Genet A 2003; 122A:125-32. PMID: 12955764
- Vitali R, Mancini C, Cesi V, Tanno B, Mancuso M, Bossi G, Zhang Y, Martinez RV, Calabretta B, Dominici C, Raschella G. Slug (SNAI2) down-regulation by RNA interference facilitates apoptosis and inhibits invasive growth in neuroblastoma preclinical models. Clin Cancer Res 2008; 14:4622-30. PMID: 18628477