GENOMIC
Mapping
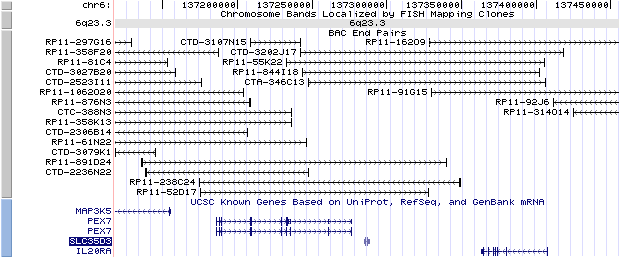
6q23.3. View the map and BAC clones (data from UCSC genome browser).
Structure
(assembly 03/2006)
SLC35D3/NM_001008783: 2 exons, 3,373 bp, chr6:137285095-137288467.
The figure below shows the structure of the SLC35D3 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Slc35d3, seq2=human SLC35D3) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
SLC35D3 (NM_001008783), 2,371bp, view ORF and the alignment to genomic.
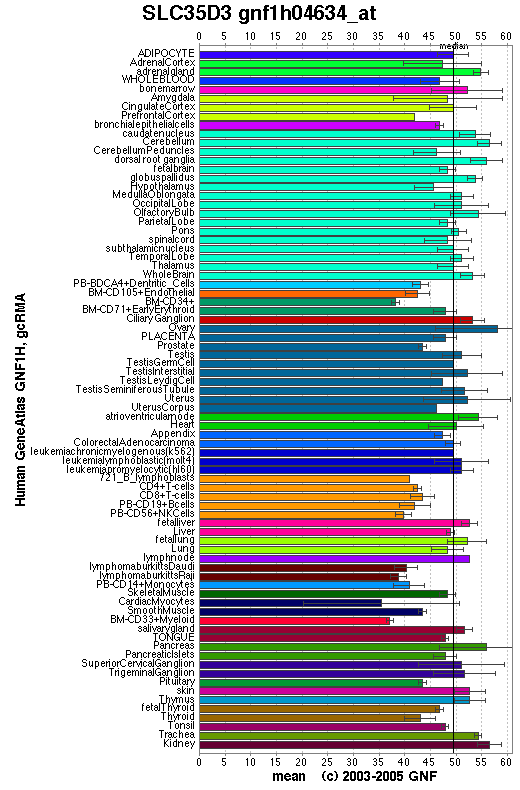
Expression Pattern
Tissue specificity:
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
solute carrier family 35, member D3 (NP_001008783): 416 aa, UniProtKB/Swiss-Prot entry Q5M8T2.
Ortholog
| Species | Chimpanzee | Dog | Mouse | Rat | Fowl |
| GeneView | SLC35D3 | LOC484001 | Slc35d3 | Slc35d3 | LOC428614 |
| Protein | XP_518764.2 (416 aa) | XP_541118.2 (423 aa) | NP_083805.1 (422 aa) | XP_218777.3 (420 aa) | XP_426171.1 (599 aa) |
| Identities | 414/416 (99%) | 380/423 (89%) | 380/422 (90%) | 388/420 (92%) | 169/245 (68%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.View evolutionary tree by TreeView
Domain
(1) Domains predicted by SMART:
(a) UAA: 13 - 305
(b) TPT: 157 - 299
(c) low complexity: 339 - 356
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a MEMBRANE PROTEIN which have 10 transmembrane helices.
| No. | N terminal | transmembrane region | C terminal | type | length |
| 1 | 9 | VLGISVAIAHGVFSGSLNILLKF | 31 | SECONDARY | 23 |
| 2 | 38 | FSFLTLVQCLTSSTAALSLELL | 59 | SECONDARY | 22 |
| 3 | 67 | VPPFGLSLARSFAGVAVLSTLQS | 89 | SECONDARY | 23 |
| 4 | 101 | LPMYVVFKRCLPLVTMLIGVLVL | 123 | PRIMARY | 23 |
| 5 | 131 | GVLAAVLITTCGAALAGAGDLTG | 153 | SECONDARY | 23 |
| 6 | 157 | GYVTGVLAVLVHAAYLVLIQKAS | 179 | PRIMARY | 23 |
| 7 | 187 | LTAQYVIAVSATPLLVICSFAST | 209 | PRIMARY | 23 |
| 8 | 224 | AMVCIFVACILIGCAMNFT | 242 | PRIMARY | 19 |
| 9 | 253 | VTTSFVGVVKSIATITVGMVAFS | 275 | SECONDARY | 23 |
| 10 | 282 | LFIAGVVVNTLGSIIYCVAKFME | 304 | SECONDARY | 23 |
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RKQS.
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRK; 366 to 368 SPR; 381 to 383 SRR; 384 to 386 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 359 to 362 SRQE; 376 to 379 SSEE; 384 to 387 SLKD.
e) N-myristoylation site: 11 to 16 GISVAI; 19 to 24 GVFSGS; 71 to 76 GLSLAR; 131 to 136 GVLAAV; 142 to 147 GAALAG; 236 to 241 GCAMNF; 259 to 264 GVVKSI; 286 to 291 GVVVNT; 342 to 347 GNGRSE; 348 to 353 GGEAAG; 365 to 370 GSPRGV; 380 to 385 GSRRSL.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1 to 52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals:
XXRR-like motif in the N-terminus: RQLC
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted comparative 3D structure of Q9WTR6 from UCSC Genome Sorter.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,182Da, pI=6.94 (NP_001008783).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer.
Pathway
SLC35D3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Screening of 22 patients with symptoms of Hermansky-Pudlak syndrome (albinism and prolonged bleeding) in which no known HPS-causing gene was found defective did not yield any SLC35D3 mutations, as did screening of 3 patients with unclassified storage pool deficiency (The latter patients have low levels of important low molecular weight components within platelet dense granules, exhibit prolonged bleeding and have no other obvious clinical symptoms.). One variation (Q317H) was identified in 2 HPS patients in the heterozygous state, which is likely not disease causing since no mutations on the other allele were identified, the Q at position 317 in SLC35D3 is not conserved in lower organisms, and one of the patients was recently identified to carry a homozygous mutation in the HPS3 gene, which explains her HPS phenotype. (Chintala, et al).
SNPs deposited in dbSNP.
Distribution
(none)
Effect
(none)
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
EDIT HISTORY:
Created by Wei Li and Min He, 12/22/2006