GENOMIC
Mapping
5q13.3. View the map and BAC clones (data from UCSC genome browser).
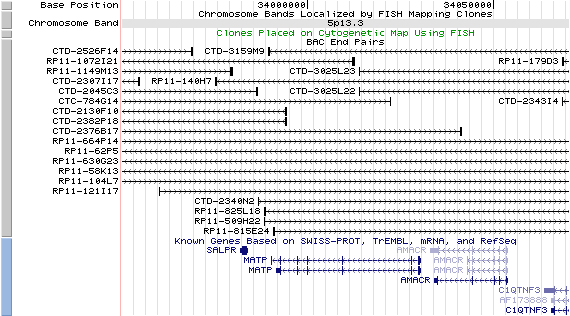
Structure
(assembly 07/03)
MATP/NM_016180: 7 exons, 40,020bp, Chr5: 33,990,231 - 34,030,250.
Note that Several alternatively spliced transcript variants has been described for this gene, but some of their full length nature has not been determined.
The figure below shows the structure of the MATP gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
MATP (NM_016180), 1,686bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: Expressed in most melanoma cell lines and melanocytes. Northern blot analysis detected 2 transcripts, one of 1.7 kb and the other of 2.8 kb (Harada, et al). As shown in the figure below, MATP may be expressed in most of the tissues examined.
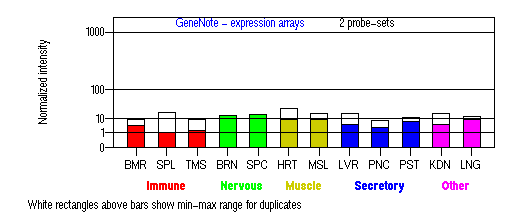
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle;
LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
Membrane-associated transporter protein (NP_057264): 530aa, ExPaSy NiceProt view of Swiss-Prot:Q9UMX9.
Synonyms: Solute carrier family 45, Member 2 (SLC45A2); AIM-1 (abscent in melanoma-1) protein; Melanoma antigen AIM1.
Ortholog
| Species | Mouse | Rat | Zebrafish | Mosquito | Fruitfly |
| GeneView | Mtap/uw | LOC310152 | 04302 | 1271101 | CG4484 |
| Protein | NP_444307 (530aa) | XP_226834 (606aa) | 014075 (554aa) | XP_309847 (521aa) | Q9VSV1 (599aa) |
| Identities | 82% /436aa | 79% /439aa | 35% /88aa | 31% /155aa | 30% /165aa |
View multiple sequence alignment (PDF files) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) transmembrane: 43 - 65
b) transmembrane: 70 - 92
c) transmembrane: 105 - 127
d) transmembrane: 137 - 159
e) transmembrane: 179 - 200
f) transmembrane: 215 - 237
g) transmembrane: 320 - 342
h) transmembrane: 365 - 387
i) transmembrane: 394 - 416
j) transmembrane: 421 - 443
k) transmembrane: 476 - 498
l) transmembrane: 503 - 525
(2) Transmembrane domains predicted by SOSUI: 10 transmembrane helices detected.
| No. | N terminal | transmembrane region | C terminal | type | length |
| 1 | 60 | LLSVGLPSSLYSIVWFLSPILGF | 82 | PRIMARY | 23 |
| 2 | 108 | TLGVMMLVGMALYLNGATVVAAL | 130 | PRIMARY | 23 |
| 3 | 138 | LVWAISVTMIGVVLFDFAADFID | 160 | PRIMARY | 23 |
| 4 | 215 | FQVMFFFSALVLTLCFTVHLCSI | 237 | PRIMARY | 23 |
| 5 | 322 | ISHLIGWTAFLSNMLFFTDFMGQ | 344 | SECONDARY | 23 |
| 6 | 370 | GCWGLCINSVFSSLYSYFQKVLV | 392 | SECONDARY | 23 |
| 7 | 398 | KGLYFTGYLLFGLGTGFIGLFPN | 420 | PRIMARY | 23 |
| 8 | 425 | LVLCSLFGVMSSTLYTVPFNLIT | 447 | PRIMARY | 23 |
| 9 | 476 | CATLTCMVQLAQILVGGGLGFLV | 498 | SECONDARY | 23 |
| 10 | 505 | VVVVITASAVALIGCCFVALFVR | 527 | PRIMARY | 23 |
Motif/Site
(1) Predicted results by ScanProsite:
a) Casein kinase II phosphorylation site : [occurs frequently]
15 - 18: SlaD,
90 - 93: SasD,
172 - 175: ShqD,
236 - 239: SisE,
254 - 257: TpqD.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site : [occurs frequently]
29 - 32: KRpT.
c) Protein kinase C phosphorylation site : [occurs frequently]
32 - 34: TsR,
270 - 272: SiK,
297 - 299: TrR,
302 - 304: TlK,
468 - 470: SvR.
d) Amidation site : [occurs frequently]
99 - 102: wGRR.
e) Cell attachment sequence : [occurs frequently]
348 - 350: RGD.
f) N-glycosylation site : [occurs frequently]
356 - 359: NSTE.
(2) Predicted results of subprograms by PSORT II:
a) Seems to no N-terminal signal peptide
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1)No ModBase entry found.
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.


2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=58,267Da, pI=7.89 (NP_057264).
FUNCTION
Ontology
a) Biological process: melanin biosynthesis from tyrosine
b) Biological process: visual perception
c) Integral to membrane
d) Belongs to the glycoside-pentoside-hexuronide (GPH) cation symporter transporter family (TC 2.A.2).
e) Melanocyte differentiation antigen
Location
Integral membrane protein; melanosome.
Interaction
Similar to the P protein, MATP have 12 transmembrane regions and are predicted to function as transporters (Costin, et al). AIM1 (absent in melanoma), a candidate suppressor of malignancy in melanoma, is a nonlens member of the betagamma-crystallin superfamily, which contains six predicted betagamma domains. The first betagamma-crystallin domain of AIM1 (AIM1-g1) diverges most in sequence from the superfamily consensus. Despite the sequence variation, AIM1-g1 folds as a betagamma domain, binds calcium and undergoes dimerization (Rajini, et al).
The Drosophila homolog, CG4484, is showing interactions in the CuraGen Drosophila interaction database.
Pathway
The MATP gene encodes a melanocyte differentiation antigen that is expressed in a high percentage of melanoma cell lines (Ray, et al). Its homolog in medaka (a small, freshwater teleost), 'B,' encodes a transporter that mediates melanin synthesis (Fukamachi, et al). Ultrastructural analysis shows that the vesicles secreted from OCA4 melanocytes are mostly early stage melanosomes. Costin, et al concluded that in OCA4 melanocytes, tyrosinase processing and intracellular trafficking to the melanosome is disrupted and the enzyme is abnormally secreted from the cells in immature melanosomes, which disrupts the normal maturation process of those organelles.
MUTATION
Allele or SNP
20 mutations of SLC45A2 deposited in HGMD.
SNPs deposited in dbSNP.
Selected allelic examples described in OMIM.
Distribution
The distribution of 30 mutations and 11 polymorphisms of SLC45A2 is described in detail in Albinism Database. 30 additional novel mutations are listed below.
(1) Hutton, et al (2008) reported 11 novel mutations, a nonsense mutation, Y278X; eight missense substitutions, G44R, H94D, G100S, R101C, S143R, G198D, M335R, A501D; and two frameshifts, c.1074delAG and c.1164delAA in Caucasian OCA4 patients.
(2) Rooryck, et al (2008)) reported five novel mutations on SLC45A2, three missense (D93N, A501D, A511E), one gross deletion (Exon 4), one nonsense (K247X) and a variant in the promoter (-1077_-1078delGA) region which was found in the heterozygous state in two of 100 controls.
(3) Li, et al (2008) reported two novel mutations (p.D160H and
c.IVS5+3delAAGT) in a Chinese OCA4 patient.
(4) Gronskov, et al (2009) reported 2 novel mutations (G416E, c.386-2A>G) in Danish OCA or AROA patients.
(5) Konno, et al (2009) reported one novel mutation (p.H38R) in a Moroccan patient.
(6) Wei, et al (2010) reported 9 previously unreported mutations:
c.-5_5delTGGCCATGGG, c.463delG, p.G110R, p.L151P, p.H233Q, p.Y266X, p.G349R, p.E368K, and p.P419L in Chinese Han OCA patients. The most common allele was p.D160H, with an allelic frequency of 55.6% in this population.
Effect
Most mutations are missense or frameshift mutations without apparent hotspots. The D157N allele might have a dominant negative effect ( Inagaki, et al(2004) ), it presents a founder effect in Japanese and Korean ( Inagaki, et al (2005) ).
PHENOTYPE
Defects in MATP (OMIM 606202) are the cause of oculocutaneous albinism type IV (OCA4) (Newton, et al) (OMIM 606574). OCA4 is characterized by hypopigmentation of skin, hair and eyes. It leads to reduced visual acuity. Two cSNPs, c.814G>A (p.Glu272Lys) and c.1122C>G (p.Phe374Leu) have shown highly significant association of MATP polymorphisms with normal human pigmentation variation (Graf, et al (2005)). In Caucasians, the -1721G, +dup (c.-1176_-1174dupAAT), and -1169A alleles in MATP were significantly associated with olive skin color. The three promoter polymorphisms were found to be in linkage disequilibrium with each other but not with the two previously reported cSNPs (Graf, et al (2007)).
REFERENCE
- Costin GE, Valencia JC, Vieira WD, Lamoreux ML, Hearing VJ. Tyrosinase processing and intracellular trafficking is disrupted in mouse primary melanocytes carrying the underwhite (uw) mutation. A model for oculocutaneous albinism (OCA) type 4. J Cell Sci 2003; 116: 3203-12. PMID: 12829739
- Fukamachi S, Shimada A, Shima A. Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nat Genet 2001; 28: 381-5. PMID: 11479596
- Graf J, Hodgson R, van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat 2005; 25: 278-84. PMID: 15714523
- Graf J, Voisey J, Hughes I, van Daal A. Promoter polymorphisms in the MATP (SLC45A2) gene are associated with normal human skin color variation. Hum Mutat 2007; 28:710-7. PMID: 17358008
- Gronskov K, Ek J, Sand A, Scheller R, Bygum A, Brixen K, Brondum-Nielsen K, Rosenberg T. Birth prevalence and mutation spectrum in Danish patients with autosomal recessive albinism. Invest Ophthalmol Vis Sci 2009; 50: 1058-64. PMID: 19060277
- Harada M, Li YF, El-Gamil M, Rosenberg SA, Robbins PF. Use of an in vitro immunoselected tumor line to identify shared melanoma antigens recognized by HLA-A*0201-restricted T cells. Cancer Res 2001; 61: 1089-94. PMID: 11221837
- Hutton SM, Spritz RA. Comprehensive Analysis of Oculocutaneous Albinism among Non-Hispanic Caucasians Shows that OCA1 Is the Most Prevalent OCA Type. J Invest Dermatol 2008; [Epub ahead of print].PMID: 18463683
- Inagaki K, Suzuki T, Shimizu H, Ishii N, Umezawa Y, Tada J, Kikuchi N, Takata M, Takamori K, Kishibe M, Tanaka M, Miyamura Y, Ito S, Tomita Y. Oculocutaneous albinism type 4 is one of the most common types of albinism in Japan. Am J Hum Genet 2004; 74: 466-71. PMID: 14961451
- Inagaki K, Suzuki T, Ito S, Suzuki N, Fukai K, Horiuchi T, Tanaka T, Manabe E, Tomita Y. OCA4: evidence for a founder effect for the p.D157N mutation of the MATP gene in Japanese and Korean. Pigment Cell Res 2005; 18: 385-8. PMID: 16162179
- Konno T, Abe Y, Kawaguchi M, Storm K, Biervliet M, Courtens W, Kono M, Tomita Y, Suzuki T. Oculocutaneous albinism type IV: A boy of Moroccan descent with a novel mutation in SLC45A2. Am J Med Genet A 2009; 149A: 1773-6.PMID: 19610114
- Li H, Meng S, Zheng H, Wei H, Zhang Y. A Chinese case of oculocutaneous albinism type 4 with two novel mutations. Int J Dermatol 2008; 47:1198ĘC201.PMID: 18986462
- Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, Brilliant MH. Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet 2001; 69: 981-8. PMID: 11574907
- Rajini B, Graham C, Wistow G, Sharma Y. Stability, homodimerization, and calcium-binding properties of a single, variant betagamma-crystallin domain of the protein absent in melanoma 1 (AIM1). Biochemistry 2003; 42: 4552-9. PMID: 12693952
- Ray ME, Wistow G, Su YA, Meltzer PS, Trent JM. AIM1, a novel non-lens member of the betagamma-crystallin superfamily, is associated with the control of tumorigenicity in human malignant melanoma. Proc Natl Acad Sci U S A 1997; 94: 3229-34. PMID: 9096375
- Rooryck C, Morice-Picard F, Elcioglu NH, Lacombe D, Taieb A, Arveiler B. Molecular diagnosis of oculocutaneous albinism: new mutations in the OCA1-4 genes and practical aspects. Pigment Cell Melanoma Res. 2008; 21: 583-7. PMID: 18821858
- Wei A, Wang Y, Long Y, Wang Y, Guo X, Zhou Z, Zhu W, Liu J, Bian X, Lian S, Li W. A comprehensive analysis reveals mutational spectra and common alleles in chinese patients with oculocutaneous albinism. J Invest Dermatol 2010; 130: 716-24. PMID: 19865097
EDIT HISTORY:
Created by Wei Li & Jonathan Bourne: 07/29/2004
Updated by Wei Li: 04/06/2006
Updated by Wei Li: 08/08/2007
Updated by Wei Li: 08/20/2008
Updated by Wei Li: 02/23/2009
Updated by Wei Li: 03/15/2010