GENOMIC
Mapping
22q12.1. View the map and BAC clones of FISH (data from UCSC genome browser).
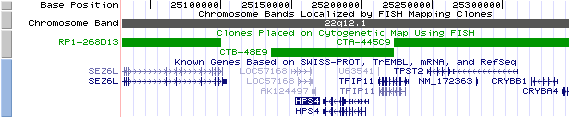
Structure
(assembly 07/03)
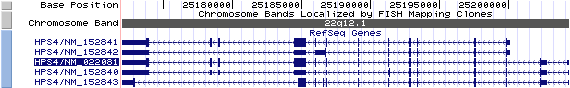
Isoform a (NM_022081): 14 exons, 32,376 bp, chr22:25,171,999-25,204,374.
Isoform b (NM_152841): 12 exons, 28,107 bp, chr22:25,171,999-25,200,105.
Isoform c (NM_152840): 14 exons, 32,376 bp, chr22:25,171,999-25,204,374.
Isoform d (NM_152843): 13 exons, 32,376 bp, chr22:25,171,999-25,204,374.
Isoform e (NM_152842): 9 exons, 28,107bp, chr22:25,171,999-25,200,105.
1) NM_152841 differs in the 5' UTR and coding region compared to NM_022081. This results in translation initiation from a downstream ATG and an isoform (b) with a distinct N-terminus compared to isoform a.
2)NM_152840 differs in the 5' UTR and includes an alternate coding segment compared to NM_022081, which causes a frameshift. The resulting isoform (c) is shorter and has a distinct C-terminus compared to isoform a.
3) NM_152843 differs in the 5' UTR and has multiple coding region differences compared to NM_022081. The resulting isoform (d) is shorter, has a distinct C-terminus, and contains an alternate aa segment compared to isoform a.
4) NM_152842 differs in the 5' UTR and has multiple coding region differences compared to NM_022081. This results in translation initiation from a downstream ATG and an isoform (e) with a shorter C-terminus and distinct N- and C-termini compared to isoform a.
The figure shows the comparison of these five isoforms (data from UCSC genome browser).
Regulatory Element
Search the 5'UTR and 1kb upstream regions of isoform a (seq1=human HPS4, seq2=mouse Hps4) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
a) Transcript variant 1 (NM_022081), 4,539bp, view ORF and the alignment to genomic.
b) Transcript variant 2 (NM_152841), 4,069bp, view ORF and the alignment to genomic.
c) Transcript variant 3 (NM_152840), 4,557bp, view ORF and the alignment to genomic.
d) Transcript variant 4 (NM_152843), 2,950bp, view ORF and the alignment to genomic.
e) Transcript variant 5 (NM_152842), 4,501bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: ubiquitous.
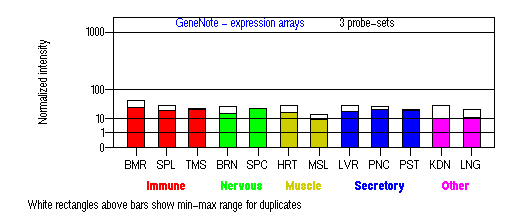
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle; LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
(HPS4p)
Isoform a ( NP_071364): 708aa, ExPaSy NiceProt view of Swiss-Prot:Q9NQG7.
Isoform b ( NP_690054): 703aa.
Isoform c ( NP_690053): 196aa.
Isoform d ( NP_690056): 528aa.
Isoform e ( NP_690055): 232aa.
Synonyms: Light-ear protein homolog.
Ortholog
| Species | Mouse | Chimpanzee | Rat | Chicken | Zebrafish | Drosophila |
| GeneView | le/Hps4 | 14194 | LOC304555 | 05595 | zgc:56538 | CG4966 |
| Protein | NP_619587 (671aa) | 24438 (708aa) | XP_222245 (545aa) | 08962 (673aa) | NP_956620 (647aa) | Q8MT61 (834aa) |
| Identities | 61%/712aa | 98%/708aa | 52%/576aa | 47%/713aa | 50%/445aa | 25%/410aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART(isoform a):
a) low complexity: 2-13
b) low complexity: 456-476
c) low complexity: 551-562
(2) Transmembrane domains predicted by SOSHI(isoform a): none.
Motif/Site
(1) Predicted results by ScanProsite(isoform a):
a) N-glycosylation site [pattern] [Warning: pattern with a high probability of occurrence]:
129 - 132 NCSQ
149 - 152 NTSD,
296 - 299 NATG,
603 - 606 NFTH,
653 - 656 NAST.
b) Protein kinase C phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
104 - 106 ScK,
210 - 212 TaK,
335 - 337 SiR,
344 - 346 SaR,
424 - 426 SlR,
465 - 467 TrR,
526 - 528 SsR,
650 - 652 TvR,
693 - 695 SgK
.
c) Amidation site [pattern]:
320 - 323 dGRK.
(2) Predicted results of subprograms by PSORT II(isoform a):
a) N-terminal signal peptide: none, N-terminal side will be inside.
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase: none.
(2) 3D models of isoform (a) predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
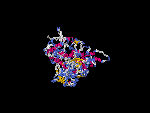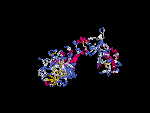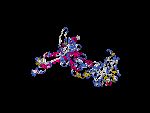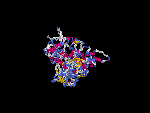
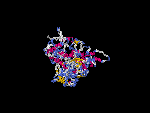
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=79,919Da, pI=5.26 (isoform a).
Computed theoretical MW=76,422Da, pI=5.30 (isoform b).
Computed theoretical MW=22,148Da, pI=4.57 (isoform c).
Computed theoretical MW=58,067Da, pI=5.34 (isoform d).
Computed theoretical MW=26,168Da, pI=5.83 (isoform e).
FUNCTION
Ontology
a) Biological process: lysosome organization and biogenesis.
b) Positive regulation of pigmentation.
c) Biogenesis of lysosome, melanosome, platelet dense granule.
Location
Cytoplasm, may be associated with membrane fraction.
Interaction
HPS4p is a component of a protein complex termed biogenesis of lysosome-related organelles complex 3 (BLOC-3), where HPS1p is residing as another subunit (Chiang, et al; Martina, et al; Nazarian, et al). The BLOC-3 complex is a moderately asymmetric complex with a molecular mass of about 175 kD (view diagram of BLOC-3 complex here). The BLOC-3 complex dissociates into smaller complex upon Tris treatment and a portion of HPS1 exists in a cytosolic complex that does not contain HPS4 (Chiang, et al). Two regions in HPS1, spanning amino acids 1-249 and 506-700 are required for binding to HPS4 (residues 340-528)(Carmona-Rivera, et al (2013)). Recombinant HPS1-HPS4 produced in insect cells can be efficiently isolated as a 1:1 heterodimer. Analytical ultracentrifugation reveals that this complex has a molecular mass of 146 kDa. BLOC-3 interacts with the GTP-bound form of the endosomal GTPase, Rab9. This interaction is mediated by HPS4 and the switch I and II regions of Rab9 (Kloer, et al). BLOC-3 is a Rab32 and Rab38 guanine nucleotide exchange factor (GEF), to promote specific membrane recruitment of Rab32/38 (Gerondopoulos, et al).
No interactions found in the CuraGen database by searching its drosophila homolog CG4966.
Pathway
HPS4 may be involved in the biogenesis of early melanosomes and the maturation or structure of cytoplasmic organelles, i.e. melanosomes, platelet dense bodies, lysosomes. Suzuki, et al revealed that HPS4 and HPS1 proteins function in the same pathway of organelle biogenesis. However, the exact mechanism of BLOC-3 in melanosomal biogenesis is lacking. Martina, et al found that HPS1 and HPS4 are components of a cytosolic complex that is involved in the biogenesis of lysosomal-related organelles through a mechanism distinct from that operated by the AP-3 complex (view diagram of BLOC-3 and AP-3 pathway here). BLOC-3 defines a novel Rab GEF family with a specific function in the biogenesis of lysosome-related organelles (Gerondopoulos, et al).
In mutant cells lacking BLOC-3, the percentages of cells displaying pronounced perinuclear accumulation were reduced (Nazarian, et al; Falcon-Perez, et al). A relatively lower frequency of microtubule-dependent movement events, either toward or away from the perinuclear region were observed in BLOC-3 deficient cells. This suggests that BLOC-3 is required for optimal attachment of late endosomes to microtubule-dependent motors (Falcon-Perez, et al).
MUTATION
Allele or SNP
9 mutations deposited in HGMD.
SNPs deposited in dbSNP.
9 selected allelic examples described in OMIM.
Distribution
| Location | Genomic | cDNA | Protein | Type | Ethnicity | Reference |
| Exon 3 | 45G>A | 45G>A | W15X | nonsense | Uruguayan | Carmona-Rivera, et al |
| Exon 3 | 47delA | 47delA | N16delA | frame-shift 26X | Uruguayan | Carmona-Rivera, et al |
| Exon 3 | 55delT | 55delT | F19delT | frame-shift 26X | European | Suzuki, et al |
| Exon 6 | 412G>T | 412G>T | E138X | nonsense | Indian | Anderson, et al |
| Exon 6 | 461A>G | 461C>T | H154R | missense | English Polish | Anderson, et al |
| Exon 7 | 541C>T | 541C>T | Q181X | nonsense | Italian | Suzuki, et al |
| Intron 7 | 597-2A>T | 597-2A>T | 597-2A>T | splicing | European | Jones, et al |
| Exon 8 | 649C>T | 649C>T | R217X | nonsense | Croation German | Anderson, et al |
| Exon 8 | 664G>T | 664G>T | E222X | nonsense | Indian | Anderson, et al |
| Exon 11 | 949-972dup | 949-972dup | A317-E324 dup 24bp | in-frame | Dutch | Suzuki, et al |
| Exon 13 | 1891C>T | 1891C>T | Q631X | nonsense | German | Suzuki, et al |
| Exon 14 | 2053delC | 2053delC | P685delC | frame-shift 701X | Sri Lanka | Bachli, et al |
| Exon 14 | 2093ins AAGCA | 2093ins AAGCA | Q698ins AAGCA | frame-shift 703X | German | Suzuki, et al |
(Numbering of genomic and cDNA sequence is based on the start codon of RefSeq NM_022081.)
No hot spot mutation is apparent.
Effect
Most of the HPS4 gene mutations are nonsense or frame-shift mutations. H154 is conserved in different species (refer to the above mutiple sequence alignment).
Most of the HPS4 gene mutations are nonsense or frame-shift mutations. H154 is conserved in different species (refer to the above mutiple sequence alignment).
PHENOTYPE
The mutation of HPS4 gene causes Hermansky-Pudlak syndrome type 4 (HPS-4, OMIM 606682 ). Anderson, et al described that HPS-4 patients exhibite a severe phenotype similar to that of patients with HPS1: iris transillumination, variable hair and skin pigmentation, absent platelet dense bodies, and occasional pulmonary fibrosis and granulomatous colitis. The patient reported from Sri Lanka (Bachli, et al) had severe pulmonary fibrosis, typical of HPS-1 disease, without granulomatous colitis. HPS4 is one of the most common gene mutated in non-Puerto Rican population.
REFERENCE
- Anderson PD, Huizing M, Claassen DA, White J, Gahl WA. Hermansky-Pudlak syndrome type 4 (HPS-4): clinical and molecular characteristics. Hum Genet 2003; 113: 10-17. PMID: 12664304
- Bachli EB, Brack T, Eppler E, Stallmach T, Trueb RM, Huizing M, Gahl WA. Hermansky-Pudlak syndrome type 4 in a patient from Sri Lanka with pulmonary fibrosis. Am J Med Genet 2004; 127A: 201-7. PMID: 15108212
- Carmona-Rivera C, Golas G, Hess RA, Cardillo ND, Martin EH, O'Brien K, Tsilou E, Gochuico BR, White JG, Huizing M, Gahl WA. Clinical, molecular, and cellular features of non-Puerto Rican Hermansky-Pudlak syndrome patients of Hispanic descent. J Invest Dermatol 2011; 131: 2394-400. PMID: 21833017
- Carmona-Rivera C, Simeonov DR, Cardillo ND, Gahl WA, Cadilla CL. A divalent interaction between HPS1 and HPS4 is required for the formation of the biogenesis of lysosome-related organelle complex-3 (BLOC-3). Biochim Biophys Acta 2013; 1833: 468-78. PMID: 23103514
- Chiang PW, Oiso N, Gautam R, Suzuki T, Swank RT, Spritz RA. The Hermansky-Pudlak syndrome 1 (HPS1) and HPS4 proteins are components of two complexes, BLOC-3 and BLOC-4, involved in the biogenesis of lysosome-related organelles. J Biol Chem 2003; 278: 20332-7. PMID: 12663659
- Falcon-Perez JM, Nazarian R, Sabatti C, Dell'angelica EC. Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J Cell Sci 2005; 118: 5243-55.PMID: 16249233
- Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol 2012; 22: 2135-9. PMID: 23084991
- Jones ML, Murden SL, Bem D, Mundell SJ, Gissen P, Daly ME, Watson SP, Mumford AD; UK GAPP study group. Rapid genetic diagnosis of heritable platelet function disorders with next-generation sequencing: proof-of-principle with Hermansky-Pudlak syndrome. J Thromb Haemost 2012; 10: 306-9. PMID: 22118648
- Kloer DP, Rojas R, Ivan V, Moriyama K, van Vlijmen T, Murthy N, Ghirlando R, van der Sluijs P, Hurley JH, Bonifacino JS. Assembly of the biogenesis of lysosome-related organelles complex-3 (BLOC-3) and its interaction with Rab9. J Biol Chem 2010; 285: 7794-804.PMID: 20048159
- Martina JA, Moriyama K, Bonifacino JS. BLOC-3, a protein complex containing the Hermansky-Pudlak syndrome gene products HPS1 and HPS4. J Biol Chem 2003; 278: 29376-84. PMID: 12756248
- Nazarian R, Falcon-Perez JM, Dell'Angelica EC. Biogenesis of lysosome-related organelles complex 3 (BLOC-3): a complex containing the Hermansky-Pudlak syndrome (HPS) proteins HPS1 and HPS4. Proc Natl Acad Sci USA 2003; 100: 8770-5. PMID: 12847290
- Suzuki T, Li W, Zhang Q, Karim A, Novak EK, Sviderskaya EV, Hill SP, Bennett DC, Levin AV, Nieuwenhuis HK, Fong CT, Castellan C, Miterski B, Swank RT, Spritz RA. Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat Genet 2002; 30: 321-4. PMID: 11836498
EDIT HISTORY:
Created by Wei Li: 06/15/2004
Updated by Wei Li: 11/24/2005
Updated by Wei Li: 06/28/2012
Updated by Wei Li: 06/13/2013