CONREAL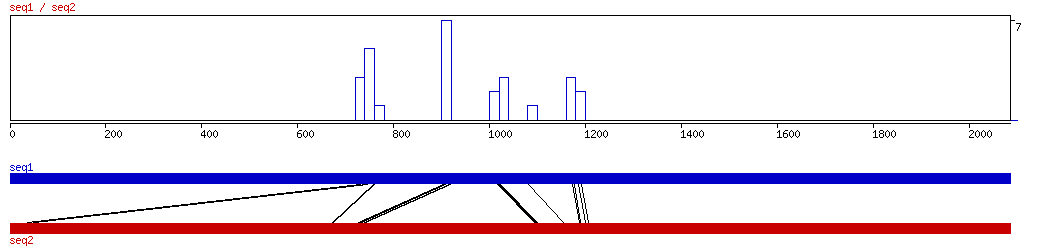
CONREAL ALIGNMENT 10 20 30 40 50 60 70 80 90 100
seq1 aagaacaaacgaaagacaacttccccaaaccaagcagatttcgggaatcactacaggacatttgtgtgaacagtcggagtattcaaagtccaggttgata
| | | | | |
seq2 ccagaatggatcatttagggccttgtaggcaatgg-----------------------------------------------------------------
10 20 30
110 120 130 140 150 160 170 180 190 200
seq1 cagaaccttctctcctaggttccagatcccaggttgtcatgggatatgttagtttcttcgttgagtaagaaataaacagctttatgtgagacacaactct
seq2 ----------------------------------------------------------------------------------------------------
210 220 230 240 250 260 270 280 290 300
seq1 gaactaggactgcccctcccagcggtcagctggggctgatactgtccaggcgtcgcccaagggaaaaggttgtggcaaggtcgctcagaaaacgcgggag
seq2 ----------------------------------------------------------------------------------------------------
310 320 330 340 350 360 370 380 390 400
seq1 gagtctggcgttgattgatgctgggaagggaccgaatgaataaaagtacttgtctggggacagcaaagaccccgctcaagctgctgaggatccgctgcct
seq2 ----------------------------------------------------------------------------------------------------
410 420 430 440 450 460 470 480 490 500
seq1 ggcattctctcggtcttttgtcagagccagagctgcattcagttgcgagaggcctgtttaggagcagctggacgcctgctgagagccgcaagctccctag
seq2 ----------------------------------------------------------------------------------------------------
510 520 530 540 550 560 570 580 590 600
seq1 ggcagttgacaacctgggaaggaggttccctgctcgtggcgctgctcctggtgcttccaatccgcacgaggtaacgaaccatcctggtttagctttgaat
seq2 ----------------------------------------------------------------------------------------------------
610 620 630 640 650 660 670 680 690 700
seq1 taacatctgtggctgaaccgtgctttggggcttccgctttgcttagtggttcgggggttttttaattagtaaaacttacacacgacagcgatttaaaccg
seq2 ----------------------------------------------------------------------------------------------------
710 720 730 740 750
seq1 agattgggttgtaaggcgagtgtttgaagtCACTGGGAcagcACAGAAGTTGGtgc--------------------------------------------
|| || | || | | | ||||||
seq2 ------------------------------CAATGAGTtagaAATGTATTTGGtgtttatcttccaaattatatttggaaatatatatgaactttattca
40 50 60 70 80 90 100
seq1 ----------------------------------------------------------------------------------------------------
seq2 gctactgggagacattggaaagtcttaagcaggggaatgacttgaattgacttagacttgcatttggtcttgctggctagggtgtggagaattgactata
110 120 130 140 150 160 170 180 190 200
seq1 ----------------------------------------------------------------------------------------------------
seq2 gacatagcaagagtgcagggagacatctcagaacatcctctgtctctcctggttatgcagacttggacatttctcggataaggcacgaacattccatctt
210 220 230 240 250 260 270 280 290 300
seq1 ----------------------------------------------------------------------------------------------------
seq2 agcagttcactgtctgccttttcctcatctatcttgcctaactcaaacctaagactctgccacctttctgctgcatcaacctaagctgctccctgctaat
310 320 330 340 350 360 370 380 390 400
seq1 ----------------------------------------------------------------------------------------------------
seq2 ggagagcaatccatacacccacagctgacttacatttattggagtccatttaagggaggataccacccctcatattgtcttatgcccaatttctgcctct
410 420 430 440 450 460 470 480 490 500
seq1 ----------------------------------------------------------------------------------------------------
seq2 gaagaaagaataagtaaaaactaaaaggcagaaatgaaatccactggcagacagtctggtgccacaccctgggtctggtagttaaagatcgacccctgac
510 520 530 540 550 560 570 580 590 600
760 770 780 790
seq1 ------------------------------------------------------------------TGATCTGTTATtctctcggttcttggctgagtgc
|| |||||| | ||| | | | |
seq2 ctaaccggttatgttatctatagattccagacattgtatggaaaagcactgtgaaaattcctgtccTGTTCTGTTCTgatctgattaccagtgcatgcag
610 620 630 640 650 660 670 680 690 700
800 810 820 830 840 850 860 870 880 890
seq1 attgacgtatgcccttggccgctaggtgccttcactccgggtccccaagcttagctaaaattgggcttgcttcttcccggcacatacgagtgttctttca
|| || |
seq2 cccccagttatgtacctgctg-------------------------------------------------------------------------------
710 720
900 910 920 930 940 950 960 970 980 990
seq1 ttgggacacgcacCCTCCCCAGTCTGGAACACCCCCtgcccgaatgctcctccccaccgactggtctacggcagggctggggattaaaactctgcgagca
| | | || || ||| ||| | | || | | || | | | | |||| | || |||
seq2 -------------CTTGCTCAATCAATCACAACCCTttcacgcagacccccttagagctgtgagcccttaaaagggataggaattgctcactcagagagc
730 740 750 760 770 780 790 800 810
1000 1010
seq1 gggtccgggaggagactgaatgcaga--------------------------------------------------------------------------
| || |
seq2 tcagctcttgagacaggagtcttgccgatgcttctggccgaataaacctcttctttctttaattcggtatccgaggaattttgtctgcagcttgtcctgc
820 830 840 850 860 870 880 890 900 910
seq1 ----------------------------------------------------------------------------------------------------
seq2 tacatttcctggttccctgactgggaagtgaggtgattggtggatggtcgaggcagctccttaggtgacttaagcctgccctgtggaacattccggtggg
920 930 940 950 960 970 980 990 1000 1010
1020 1030
seq1 --------------------------------------------------------------------------------CCAGCGGGTGGCGTgcgccc
||| | ||||| || |||
seq2 ggactctggccagcccgagcaacgtggatcctgagagcactcccaggtaggcatttgccccggtgggacgccttgccagaGCAGTGTGTGGCAGgccccc
1020 1030 1040 1050 1060 1070 1080 1090 1100 1110
1040 1050 1060 1070 1080 1090 1100 1110 1120 1130
seq1 aagttccccactggcgcgcaaacttaacttactgttaaggcgcggGTAGAGGcaaccgggctagtgtgttttcagaggcttggctggggtagctgactta
| | ||| || ||| | | | || | | |
seq2 gtggaggatcaacacagtggctgaacactgggaaggaactg----GTACTTGgagtctggacatctgaaacttggct-----------------------
1120 1130 1140 1150 1160 1170 1180
1140 1150 1160 1170 1180 1190 1200 1210 1220
seq1 agtgtcctgtcttcactccccagttggtctccagacTGAAAACAGCagAGCGGCtACCAGActctcacaggagcaagctgtaac-
|||| || ||||||| ||| ||| | | ||
seq2 ------------------------------------CTGAAACTGCggAGCGGCcACCGGAcgccttctgg-------------A
1190 1200 1210 1220
CONREAL ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00314* | 50.00 | 731 | 738 | 36 | 43 | 2 | 4.747 | 5.823 | 0.82 | 0.87 | Pro only | C only | | M00313* | 50.00 | 731 | 738 | 36 | 43 | 2 | 4.434 | 5.631 | 0.81 | 0.87 | Pro only | C only | | M00315* | 50.00 | 731 | 738 | 36 | 43 | 2 | 4.504 | 5.763 | 0.81 | 0.87 | Pro only | C only | | M00630* | 58.97 | 743 | 751 | 48 | 56 | 2 | 5.430 | 6.739 | 0.81 | 0.86 | Pro only | C only | | M00396 | 59.46 | 746 | 752 | 51 | 57 | 1 | 5.185 | 6.399 | 0.89 | 0.96 | En-1 | C only | | M00640* | 57.89 | 746 | 753 | 51 | 58 | 1 | 5.193 | 5.661 | 0.80 | 0.82 | Pro only | C only | | M00253 | 50.00 | 757 | 764 | 672 | 679 | 2 | 3.360 | 3.262 | 0.83 | 0.83 | cap | C only | | M00747* | 51.35 | 759 | 765 | 674 | 680 | 1 | 5.830 | 8.757 | 0.81 | 0.92 | Pro only | C only | | M00921* | 52.63 | 760 | 767 | 675 | 682 | 1 | 5.500 | 10.200 | 0.82 | 1.00 | Pro only | C only | | M00658* | 50.00 | 904 | 911 | 727 | 734 | 2 | 5.780 | 7.116 | 0.80 | 0.85 | Pro only | C only | | M00314* | 52.63 | 907 | 914 | 730 | 737 | 1 | 7.015 | 6.538 | 0.93 | 0.91 | Pro only | C only | | M00313* | 52.63 | 907 | 914 | 730 | 737 | 1 | 6.815 | 6.341 | 0.92 | 0.90 | Pro only | C only | | M00315* | 52.63 | 907 | 914 | 730 | 737 | 1 | 7.134 | 6.221 | 0.94 | 0.90 | Pro only | C only | | M00253 | 55.26 | 909 | 916 | 732 | 739 | 1 | 5.123 | 3.337 | 0.91 | 0.83 | cap | C only | | M01020* | 52.50 | 917 | 926 | 740 | 749 | 2 | 9.064 | 5.945 | 0.90 | 0.80 | Pro only | C only | | M00253 | 55.26 | 919 | 926 | 742 | 749 | 1 | 4.718 | 3.296 | 0.89 | 0.83 | cap | C only | | M00253 | 52.63 | 1017 | 1024 | 1094 | 1101 | 1 | 2.964 | 4.328 | 0.82 | 0.87 | cap | C L M | | M00468* | 51.35 | 1018 | 1024 | 1095 | 1101 | 1 | 6.741 | 7.069 | 0.91 | 0.93 | Pro only | C L M | | M00253 | 50.00 | 1020 | 1027 | 1097 | 1104 | 2 | 4.482 | 3.546 | 0.88 | 0.84 | cap | C L M | | MA0006 | 52.78 | 1021 | 1026 | 1098 | 1103 | 1 | 4.891 | 6.158 | 0.80 | 0.85 | Ahr-ARNT | C L M | | M00428* | 50.00 | 1023 | 1030 | 1100 | 1107 | 1 | 5.430 | 3.797 | 0.86 | 0.80 | Pro only | C L M | | M00396 | 51.35 | 1082 | 1088 | 1155 | 1161 | 1 | 4.052 | 5.759 | 0.83 | 0.92 | En-1 | C L M | | M00415 | 53.85 | 1173 | 1181 | 1187 | 1195 | 2 | 5.779 | 7.297 | 0.83 | 0.89 | AREB6 | C only | | M00486* | 53.85 | 1173 | 1181 | 1187 | 1195 | 1 | 4.083 | 6.843 | 0.80 | 0.92 | Pro only | C only | | M00148 | 56.76 | 1176 | 1182 | 1190 | 1196 | 1 | 6.390 | 5.665 | 0.89 | 0.86 | SRY | C L M | | M00803* | 61.11 | 1185 | 1190 | 1199 | 1204 | 2 | 4.573 | 4.573 | 0.80 | 0.80 | Pro only | C L M | | M00271 | 72.22 | 1192 | 1197 | 1206 | 1211 | 2 | 6.449 | 5.085 | 0.86 | 0.82 | AML-1a | C L M |
LAGAN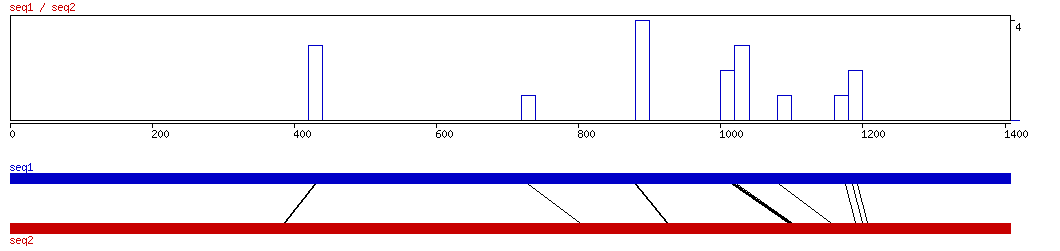
LAGAN ALIGNMENT 10 20 30 40 50 60 70 80 90 100
seq1 aagaacaaacgaaagacaacttccccaaaccaagcagatttcgggaatcactacaggacatttgtgtgaacagtcggagtattcaaagtccaggttgata
| | ||| | | ||| || ||| ||||| || | ||
seq2 c----------------------------cagaatggatcatttagggccttgtaggcaatggcaatgagttagaaatgtatttggtgtttatctt----
10 20 30 40 50 60
110 120 130 140 150 160 170 180 190
seq1 cagaaccttctctcctaggttccagatcccaggttgtcatgggatatgtt-----agtttcttcgttgagtaagaaata---aacagctttatgtgagac
||| || | | ||| || | | ||| ||| | | || | | | || ||| | |
seq2 ----------------------------ccaaattatatttggaaatatatatgaactttattcagctactgggagacattggaaagtcttaagcagggg
70 80 90 100 110 120 130 140
200 210 220 230 240 250 260 270 280 290
seq1 acaactctgaactaggactgcccctcccagcggtcagctggggctgatactgtccaggcgtcgcccaagggaaaaggttgtggcaaggtcgctcagaaaa
| |||| | | | | | ||||| | | | | | ||| || | || ||||||| |
seq2 aatgacttgaattgacttagacttgcatttggtcttgctggctagggtgtggagaattgactatagacatagcaagagtgcagggagacatctcagaaca
150 160 170 180 190 200 210 220 230 240
300 310 320 330 340 350 360 370 380
seq1 ----------cgcgggaggagtctggcgttgattgatgctgggaagggaccgaatgaataaaagtacttgtctggggacagcaaagaccccgctcaagct
| | | | | | | | || | || | || |||| | || || | | || || | | |
seq2 tcctctgtctctcctggttatgcagacttggacatttctcggataaggcacgaacattccatcttagcagttcactgtctgccttttcctcatctatctt
250 260 270 280 290 300 310 320 330 340
390 400 410 420 430 440 450 460 470 480
seq1 gctgaggatccgctgcctggcattctctcggtcttttgtcagagcCAGAGCTGCAttcagttgcgagaggcctgtttaggagcagctggacgcctgctga
|| | | | | | ||| || | || |||||| | | | ||| | | | | | | ||
seq2 gcctaactcaaacctaagactctgccacctttctgctgcatcaacCTAAGCTGCTccctgctaatggagagcaatccatacacccacagctgacttacat
350 360 370 380 390 400 410 420 430 440
490 500 510 520
seq1 gagccgcaagctccctagggcagttgacaacct-------------------------------------------------------------gggaag
| | | || || || || ||| | ||
seq2 ttattggagtccatttaagggaggataccacccctcatattgtcttatgcccaatttctgcctctgaagaaagaataagtaaaaactaaaaggcagaaat
450 460 470 480 490 500 510 520 530 540
530 540 550 560 570 580 590 600 610
seq1 gaggttccctgctcg------tggcgctgctcctggtgcttccaatccgcacgaggtaacgaaccatcctggtttagctttgaattaacatctgtggct-
|| | | ||| | ||| || | || | | | || ||| || | | || | | | |||| | | |
seq2 gaaatccactggcagacagtctggtgccacaccctgggtct---------ggtagttaaagatcgacccctgacctaaccggttatgttatctatagatt
550 560 570 580 590 600 610 620 630
620 630 640 650 660
seq1 --------------gaaccgtgctttggggcttcc----------------gctttgcttagtggttcgggggttttttaatta----------------
||| | || || |||| | | || ||| || | | | |||
seq2 ccagacattgtatggaaaagcactgtgaaaattcctgtcctgttctgttctgatctgattaccagtgcatgcagcccccagttatgtacctgctgcttgc
640 650 660 670 680 690 700 710 720 730
670 680 690 700 710 720 730 740 750 7
seq1 ----------gtaaaacttacacacgacagcgatttaaaccgagattgggttgtaaggcgagtgtttgaagTCACTGGGacagcacagaagttggtgctg
|| ||| ||| | | || | | | || | | || || ||| ||||| || |||
seq2 tcaatcaatcacaaccctttcacgcagacccccttagagctgtgagcccttaaaagggataggaattgc--TCACTCAGagagc----------------
740 750 760 770 780 790 800 810
60 770 780 790 800 810 820 830 840 850
seq1 atctgttattctctcggttcttggctgagtgcattgacgtatgcccttggccgc-taggtgccttcactccgggtccccaagcttagctaaaattgggct
|| | ||||| || | | |||| |||||| || |||| || | || |||
seq2 -------------tcagctcttgagacaggagtcttgccgatgcttctggccgaataaacctcttctttctttaattcggtatccgaggaattttgtctg
820 830 840 850 860 870 880 890 900
860 870 880 890 900 910 920 930
seq1 tgcttcttcccggcacatacgaGTGTTCTTtcattgggacacgca-------------------------ccctccccagtctggaacaccccctgcccg
| ||| | |||| |||| | | ||||| | ||| | || | | |||||
seq2 cagcttgtcctgctacatttccTGGTTCCCtgactgggaagtgaggtgattggtggatggtcgaggcagctccttaggtgacttaagcctgccctg----
910 920 930 940 950 960 970 980 990
940 950 960 970 980 990 1000 1010 1
seq1 aatgctcctccccaccgactggtctacggcagggctggggattaaaactctgcgagcagggtccgggaggagactgaatg---------------cagaC
|| | || | | | ||| | | | | ||| ||||| | || ||| || ||||
seq2 --tggaacattccggtgggggactctggccagcccgagcaacgtggatcctgagagcactcccaggtaggcatttgccccggtgggacgccttgccagaG
1000 1010 1020 1030 1040 1050 1060 1070 1080 1090
020 1030 1040 1050 1060 1070 1080 1090 1100 1110
seq1 CAGCGGGTGGCGTgcgccc----aagttccccactggcgcgcaaacttaacttactgttaaggcgcggGTAGAGGcaaccgggctagtgtgttttcagag
||| | ||||| || ||| | | || ||| | || || |||| |||| | |||| | | | || | || ||
seq2 CAGTGTGTGGCAGgcccccgtggaggatcaacacagtggctgaa--------cactgggaaggaactgGTACTTGgagtctggaca--------tctgaa
1100 1110 1120 1130 1140 1150 1160 1170 11
1120 1130 1140 1150 1160 1170 1180 1190 1200 1210
seq1 gcttggctggggtagctgacttaagtgtcctgtcttcactccccagttggtctccagactgaAAACAGCagAGCGGCtACCAGActctcacaggagcaag
||||||| | |||| || ||||||| ||| ||| | | |||||| |
seq2 acttggctctg---------------------------------------------------AAACTGCggAGCGGCcACCGGAcgccttctggagcagg
80 1190 1200 1210 1220 1
1220
seq1 ctgtaac
| | |
seq2 tagcagc
230
LAGAN ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00176 | 50.00 | 428 | 437 | 386 | 395 | 1 | 6.879 | 7.594 | 0.81 | 0.84 | AP-4 | L only | | M00253 | 52.63 | 429 | 436 | 387 | 394 | 2 | 5.063 | 4.332 | 0.90 | 0.87 | cap | L only | | M00644* | 51.35 | 430 | 436 | 388 | 394 | 1 | 6.451 | 6.451 | 0.85 | 0.85 | Pro only | L only | | M00253 | 52.63 | 730 | 737 | 801 | 808 | 1 | 4.347 | 4.980 | 0.87 | 0.90 | cap | L M | | M00497* | 52.63 | 880 | 887 | 923 | 930 | 1 | 4.029 | 7.275 | 0.81 | 0.96 | Pro only | L M | | M00499* | 52.63 | 880 | 887 | 923 | 930 | 1 | 7.190 | 3.834 | 0.95 | 0.81 | Pro only | L M | | M00500* | 52.63 | 880 | 887 | 923 | 930 | 1 | 4.549 | 4.220 | 0.83 | 0.82 | Pro only | L M | | MA0080 | 50.00 | 882 | 887 | 925 | 930 | 2 | 4.249 | 7.727 | 0.81 | 0.96 | SPI-1 | L M | | M00253 | 52.63 | 1017 | 1024 | 1094 | 1101 | 1 | 2.964 | 4.328 | 0.82 | 0.87 | cap | L C M | | M00468* | 51.35 | 1018 | 1024 | 1095 | 1101 | 1 | 6.741 | 7.069 | 0.91 | 0.93 | Pro only | L C M | | M00253 | 50.00 | 1020 | 1027 | 1097 | 1104 | 2 | 4.482 | 3.546 | 0.88 | 0.84 | cap | L C M | | MA0006 | 52.78 | 1021 | 1026 | 1098 | 1103 | 1 | 4.891 | 6.158 | 0.80 | 0.85 | Ahr-ARNT | L C M | | M00428* | 50.00 | 1023 | 1030 | 1100 | 1107 | 1 | 5.430 | 3.797 | 0.86 | 0.80 | Pro only | L C M | | M00396 | 51.35 | 1082 | 1088 | 1155 | 1161 | 1 | 4.052 | 5.759 | 0.83 | 0.92 | En-1 | L C M | | M00148 | 56.76 | 1176 | 1182 | 1190 | 1196 | 1 | 6.390 | 5.665 | 0.89 | 0.86 | SRY | L C M | | M00803* | 61.11 | 1185 | 1190 | 1199 | 1204 | 2 | 4.573 | 4.573 | 0.80 | 0.80 | Pro only | L C M | | M00271 | 72.22 | 1192 | 1197 | 1206 | 1211 | 2 | 6.449 | 5.085 | 0.86 | 0.82 | AML-1a | L C M |
MAVID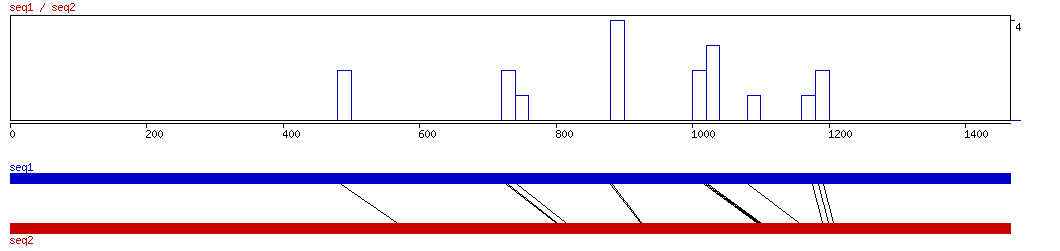
MAVID ALIGNMENT 10 20 30 40 50 60 70
seq1 -----aagaacaaacgaaagacaacttccccaaaccaagcag----------atttcgggaatcactaca-----------ggacatttgtgtgaac---
| | | | | | | | | | ||| || |||| | | | || | ||| || | | |||||
seq2 ccagaatggatcatttagggccttgtaggcaatggcaatgagttagaaatgtatttggtgtttatcttccaaattatatttggaaatatatatgaacttt
10 20 30 40 50 60 70 80 90 100
80
seq1 -------------------------agtc-------------------------------------------------------------ggagtattca
|||| |||| ||| |
seq2 attcagctactgggagacattggaaagtcttaagcaggggaatgacttgaattgacttagacttgcatttggtcttgctggctagggtgtggagaattga
110 120 130 140 150 160 170 180 190 200
90 100 110 120 130 140 150 160
seq1 a---------------agtccaggttgata---cagaaccttctctcctaggttccagatcccaggttgtcatgg-gatatgttagtttcttcgttgagt
||| |||| || | |||||| | |||| || | | || ||| || || ||||| | | ||
seq2 ctatagacatagcaagagtgcagggagacatctcagaacatcctct------------gtctctcctggttatgcagacttggacatttctcggataagg
210 220 230 240 250 260 270 280 2
170 180 190 200 210 220 230 2
seq1 aagaaata---------aacagctttatgtgagacac-------------------aactctgaactaggactgcccctcccagcggtcagctggggctg
| || | | ||| | ||| | | ||||| | ||| |||| || ||| | || ||| | |
seq2 cacgaacattccatcttagcagttcactgtctgccttttcctcatctatcttgcctaactcaaacctaagact----ctgccacctttctgct--gcatc
90 300 310 320 330 340 350 360 370 380
40 250 260 270 280 290 300 310 320 330 3
seq1 atactgtccaggcgtcgcccaagggaaaaggttgtggcaaggtcgctcagaaaacgcgggaggagtctggcgttgattgatgctgggaagggaccgaatg
| || | | | | || ||| | |||| || | || | || | || | || |||| || |
seq2 aacctaagctgctccctgctaatggaga--------gcaa------tccatacac-ccacagctgacttacatttattg----------------gagtc
390 400 410 420 430 440 450
40 350 360 370 380 390 400 410 420 430 4
seq1 aataaaagtacttgtctggggacagcaaagaccccgctcaagctgctgaggatccgctgcctggcattctctcggtcttttgtcagagccagagctgcat
|| ||| | ||| | || || |||| || || |||| || | ||| ||| | ||| ||
seq2 catttaag--------ggaggatacca------cccctcatattg-------tcttatgcc---caatttctgcctctgaagaaagaataag--------
460 470 480 490 500 510 52
40 450 460 470 480 490 500 510 520 530 5
seq1 tcagttgcgagaggcctgtttaggagcagctggacgcctgctgagaGCCGCAAGctccctagggcagttgacaacctgggaaggaggttccctgctcgtg
| | | |||| | | | |||| | | | | ||| || || | || |||| ||| | ||||| |
seq2 taaaaactaaaaggcagaaatgaaatccactggcagacagtctggtGCCACACCctgggtctggtagtt----------aaagatcgacccctgacctaa
0 530 540 550 560 570 580 590 600 61
40 550 560 570 580 590 600 610 6
seq1 gcgctgctc----ctggtgcttccaatcc------------gcacgaggtaacgaaccatcctggtttagctttga----attaacatctgtggctgaac
|| | | || | ||||| | |||| | || | |||| ||| | | ||| |||| || || | |
seq2 ccggttatgttatctatagattccagacattgtatggaaaagcactgtgaaaattcctgtcct-gttctgttctgatctgattaccagtgcatgcagccc
0 620 630 640 650 660 670 680 690 700 7
20 630 640 650 660 670 680 690 700 710 7
seq1 cgtgctttggggcttccgctttgcttagtggttcgggggttttttaattagtaaaacttacacacgacagcgatttaaaccgagattgggttgtaaggcg
| | | || || | || ||||| | | || ||| | || |||| | | ||| || ||| | ||| || |
seq2 ccagttatgtacctgctgc-ttgctcaatcaatcacaaccctttcacgcagacccccttagagctgtgagcccttaaaa--gggataggaat--------
10 720 730 740 750 760 770 780 790
20 730 740 750 760 770 780 790 800 810 8
seq1 agtgtttgaAGTCACTGGGacagcACAGAAGTtggtgctgatctgttattctctcggttcttggctgagtgcattgacgtatgcccttggccgctaggtg
||||| || ||| ||| ||| | | | | | || | ||| ||| | || || | | | |
seq2 --------tGCTCACTCAGagagcTCAGCTCTtgagacaggagtct-----tgccgatgctt--ctggccgaataaacctcttc--------------tt
800 810 820 830 840 850 860 8
20 830 840 850 860 870 880 890 900
seq1 ccttcactccgggtccccaagcttagctaaaattgggcttgcttctt--cccggcacatacgaGTGTTCTTtcattgggacacgc---------------
||| | | ||| || | | |||| | ||| ||| || | |||| |||| | | ||||| |
seq2 tctttaattcggtatccgagg---------aattttgtctgcagcttgtcctgctacatttccTGGTTCCCtgactgggaagtgaggtgattggtggatg
70 880 890 900 910 920 930 940 950 96
910 920 930 940 950 960 970
seq1 --------------------------accctccccagtctggaacaccccc-tgcccgaatgctcctccccaccgactggtctacggcagggctggggat
| ||| ||| | ||||||| || || || | |||| | | || | ||||
seq2 gtcgaggcagctccttaggtgacttaagcctgccc--tgtggaacattccggtgggggact---------------ctggccagcccgagcaacgtggat
0 970 980 990 1000 1010 1020 1030 1040
980 990 1000 1010 1020 1030 1040 1050
seq1 taaaactctgcgagcagggtccgggagga--------------gactgaatgc-agaCCAGCGGGTGGCGTgcgccc----aagttccccactggcgcgc
||| ||||| | || ||| ||| ||| ||| ||| | ||||| || ||| | | || ||| | ||
seq2 ------cctgagagcactcccaggtaggcatttgccccggtgggacgccttgccagaGCAGTGTGTGGCAGgcccccgtggaggatcaacacagtggctg
1050 1060 1070 1080 1090 1100 1110 1120 1130
1060 1070 1080 1090 1100 1110 1120 1130 1140 1150
seq1 aaacttaacttactgttaaggcgcggGTAGAGGcaaccgggctagtgtgttttcagaggcttggctggggtagctgacttaagtgtcctgtcttcactcc
|| |||| |||| | |||| | | | || | || || ||||||| |
seq2 aa--------cactgggaaggaactgGTACTTGgagtctggaca--------tctgaaacttggctctg-------------------------------
1140 1150 1160 1170 1180
1160 1170 1180 1190 1200 1210 1220
seq1 ccagttggtctccagactgaAAACAGCagAGCGGCtACCAGActctcacaggagcaagctgtaac
|||| || ||||||| ||| ||| | | |||||| | | | |
seq2 --------------------AAACTGCggAGCGGCcACCGGAcgccttctggagcaggtagcagc
1190 1200 1210 1220 1230
MAVID ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00271 | 50.00 | 485 | 490 | 566 | 571 | 2 | 5.416 | 6.779 | 0.83 | 0.88 | AML-1a | M only | | M00731* | 52.63 | 485 | 492 | 566 | 573 | 1 | 6.315 | 7.359 | 0.80 | 0.84 | Pro only | M only | | MA0067 | 52.63 | 728 | 735 | 799 | 806 | 1 | 8.226 | 4.165 | 0.97 | 0.81 | Pax-2 | M only | | M00253 | 52.63 | 730 | 737 | 801 | 808 | 1 | 4.347 | 4.980 | 0.87 | 0.90 | cap | M L | | M00253 | 55.26 | 743 | 750 | 814 | 821 | 1 | 4.009 | 6.009 | 0.86 | 0.94 | cap | M only | | M00497* | 52.63 | 880 | 887 | 923 | 930 | 1 | 4.029 | 7.275 | 0.81 | 0.96 | Pro only | M L | | M00499* | 52.63 | 880 | 887 | 923 | 930 | 1 | 7.190 | 3.834 | 0.95 | 0.81 | Pro only | M L | | M00500* | 52.63 | 880 | 887 | 923 | 930 | 1 | 4.549 | 4.220 | 0.83 | 0.82 | Pro only | M L | | MA0080 | 50.00 | 882 | 887 | 925 | 930 | 2 | 4.249 | 7.727 | 0.81 | 0.96 | SPI-1 | M L | | M00253 | 52.63 | 1017 | 1024 | 1094 | 1101 | 1 | 2.964 | 4.328 | 0.82 | 0.87 | cap | M C L | | M00468* | 51.35 | 1018 | 1024 | 1095 | 1101 | 1 | 6.741 | 7.069 | 0.91 | 0.93 | Pro only | M C L | | M00253 | 50.00 | 1020 | 1027 | 1097 | 1104 | 2 | 4.482 | 3.546 | 0.88 | 0.84 | cap | M C L | | MA0006 | 52.78 | 1021 | 1026 | 1098 | 1103 | 1 | 4.891 | 6.158 | 0.80 | 0.85 | Ahr-ARNT | M C L | | M00428* | 50.00 | 1023 | 1030 | 1100 | 1107 | 1 | 5.430 | 3.797 | 0.86 | 0.80 | Pro only | M C L | | M00396 | 51.35 | 1082 | 1088 | 1155 | 1161 | 1 | 4.052 | 5.759 | 0.83 | 0.92 | En-1 | M C L | | M00148 | 56.76 | 1176 | 1182 | 1190 | 1196 | 1 | 6.390 | 5.665 | 0.89 | 0.86 | SRY | M C L | | M00803* | 61.11 | 1185 | 1190 | 1199 | 1204 | 2 | 4.573 | 4.573 | 0.80 | 0.80 | Pro only | M C L | | M00271 | 72.22 | 1192 | 1197 | 1206 | 1211 | 2 | 6.449 | 5.085 | 0.86 | 0.82 | AML-1a | M C L |
BLASTZ
BLASTZ ALIGNMENT 10 20 30 40 50 60 70 80 90 100
seq1 aagaacaaacgaaagacaacttccccaaaccaagcagatttcgggaatcactacaggacatttgtgtgaacagtcggagtattcaaagtccaggttgata
| | | | | | || | || | ||| | | | | | | || | | | |
seq2 ccagaatggatcatttagggccttgtaggcaatggcaatgagttagaaatgtatttggtgtttatcttccaaattatatttggaaatatatatgaacttt
10 20 30 40 50 60 70 80 90 100
110 120 130 140 150 160 170 180 190 200
seq1 cagaaccttctctcctaggttccagatcccaggttgtcatgggatatgttagtttcttcgttgagtaagaaataaacagctttatgtgagacacaactct
| || || | || | | | | | | ||| | | | | || | | || |
seq2 attcagctactgggagacattggaaagtcttaagcaggggaatgacttgaattgacttagacttgcatttggtcttgctggctagggtgtggagaattga
110 120 130 140 150 160 170 180 190 200
210 220 230 240 250 260 270 280 290 300
seq1 gaactaggactgcccctcccagcggtcagctggggctgatactgtccaggcgtcgcccaagggaaaaggttgtggcaaggtcgctcagaaaacgcgggag
| | || || | | | | | | | | | | | |
seq2 ctatagacatagcaagagtgcagggagacatctcagaacatcctctgtctctcctggttatgcagacttggacatttctcggataaggcacgaacattcc
210 220 230 240 250 260 270 280 290 300
310 320 330 340 350 360 370 380 390 400
seq1 gagtctggcgttgattgatgctgggaagggaccgaatgaataaaagtacttgtctggggacagcaaagaccccgctcaagctgctgaggatccgctgcct
| | ||| | || || | | || | | | |
seq2 atcttagcagttcactgtctgccttttcctcatctatcttgcctaactcaaacctaagactctgccacctttctgctgcatcaacctaagctgctccctg
310 320 330 340 350 360 370 380 390 400
410 420 430 440 450 460 470 480 490 500
seq1 ggcattctctcggtcttttgtcagagccagagctgcattcagttgcgagaggcctgtttaggagcagctggacgcctgctgagagccgcaagctccctag
|| | | | | | ||| | | | | | | | |
seq2 ctaatggagagcaatccatacacccacagctgacttacatttattggagtccatttaagggaggataccacccctcatattgtcttatgcccaatttctg
410 420 430 440 450 460 470 480 490 500
510 520 530 540 550 560 570 580 590 600
seq1 ggcagttgacaacctgggaaggaggttccctgctcgtggcgctgctcctggtgcttccaatccgcacgaggtaacgaaccatcctggtttagctttgaat
|| | | | | | | | | | | | | | | | || |
seq2 cctctgaagaaagaataagtaaaaactaaaaggcagaaatgaaatccactggcagacagtctggtgccacaccctgggtctggtagttaaagatcgaccc
510 520 530 540 550 560 570 580 590 600
610 620 630 640 650 660 670 680 690 700
seq1 taacatctgtggctgaaccgtgctttggggcttccgctttgcttagtggttcgggggttttttaattagtaaaacttacacacgacagcgatttaaaccg
| | | || | | | ||| | | | | | | | | | | | |
seq2 ctgacctaaccggttatgttatctatagattccagacattgtatggaaaagcactgtgaaaattcctgtcctgttctgttctgatctgattaccagtgca
610 620 630 640 650 660 670 680 690 700
710 720 730 740 750 760 770 780 790 800
seq1 agattgggttgtaaggcgagtgtttgaagtcactgggacagcacagaagttggtgctgatctgttattctctcggttcttggctgagtgcattgacgtat
| | || | | | | || || || | | || | | || | || |
seq2 tgcagcccccagttatgtacctgctgcttgctcaatcaatcacaaccctttcacgcagacccccttagagctgtgagcccttaaaagggataggaattgc
710 720 730 740 750 760 770 780 790 800
810 820 830 840 850 860 870 880 890 900
seq1 gcccttggccgctaggtgccttcactccgggtccccaagcttagctaaaattgggcttgcttcttcccggcacatacgagtgttctttcattgggacacg
| || | ||| | | || | | | | | ||||| | || | | | | |
seq2 tcactcagagagctcagctcttgagacaggagtcttgccgatgcttctggccgaataaacctcttctttctttaattcggtatccgaggaattttgtctg
810 820 830 840 850 860 870 880 890 900
910 920 930 940 950 960 970 980 990 1000
seq1 caccctccccagtctggaacaccccctgcccgaatgctcctccccaccgactggtctacggcagggctggggattaaaactctgcgagcagggtccggga
|| | | || | | | || || || | | | ||| | ||| || | | |||
seq2 cagcttgtcctgctacatttcctggttccctgactgggaagtgaggtgattggtggatggtcgaggcagctccttaggtgacttaagcctgccctgtgga
910 920 930 940 950 960 970 980 990 1000
1010 1020 1030 1040 1050 1060 1070 1080 1090 1100
seq1 ggagactgaatgcagaccagcgggtggcgtgcgcccaagttccccactggcgcgcaaacttaacttactgttaaggcgcgggtagaggcaaccgggctag
| | | | ||| | | | | | || || | | | || || || | | || | | |
seq2 acattccggtgggggactctggccagcccgagcaacgtggatcctgagagcactcccaggtaggcatttgccccggtgggacgccttgccagagcagtgt
1010 1020 1030 1040 1050 1060 1070 1080 1090 1100
1110 1120 1130 1140 1150 1160 1170 1180 1190 1200
seq1 tgtgttttcagaggcttggctggggtagctgacttaagtgtcctgtcttcactccccagttggtctccagactgaaaacagcagagcggctaccagactc
| | | | | | | | ||| | | | || | | | ||
seq2 gtggcaggcccccgtggaggatcaacacagtggctgaacactgggaaggaactggtacttggagtctggacatctgaaacttggctctgaaactgcggag
1110 1120 1130 1140 1150 1160 1170 1180 1190 1200
1210 1220
seq1 tcacaggagcaagctgtaac--------------
| | | || |
seq2 cggccaccggacgccttctggagcaggtagcagc
1210 1220 1230
BLASTZ ALIGNED HITSNo hits found
Run time: 88.8 sec | 




