GENOMIC
Mapping
7qB4. View the map and BAC clones (data from UCSC genome browser, assembly 02/03).
Structure
(assembly 02/03)
P/NM_021879: 22 exons, 299,358bp, Chr7: 44,957,913 - 45,257,270.
The figure below shows the map of the P gene (data from UCSC genome browser).
Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
P (NM_021879), 3,119bp, view ORF and the alignment to genomic.
Expression Pattern
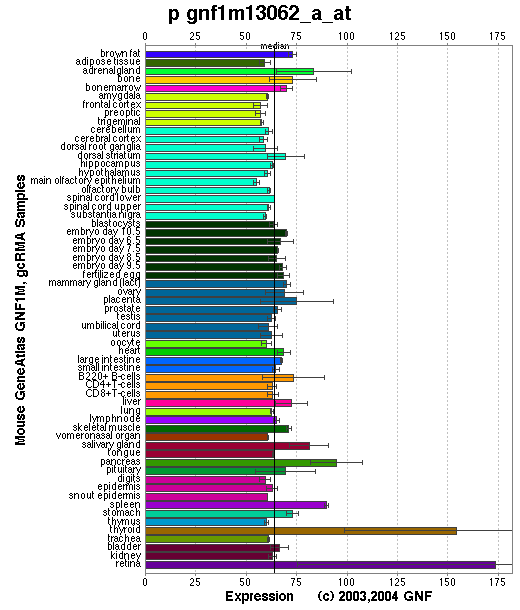
Tissue specificity: Varied expression, highest in skin and eyes.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
P protein (NP_068679): 833aa,
ExPaSy NiceProt view of Swiss-Prot:Q62052.
Ortholog
| Species | Human | Zebrafish | Fruitfly | Mosquito |
| GeneView | OCA2 | 07116 | CG15624/hoe2 | 1280250 |
| Protein | NP_000266 (838aa) | 13636 (128aa) | NP_608878 (846aa) | XP_320080 (580aa) |
| Identities | 78% /659aa | 54% /70aa | 42% /214aa | 50% /260aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) transmembrane 171 - 193
b) transmembrane 319 - 341
c) transmembrane 348 - 365
d) transmembrane 380 - 402
e) transmembrane 415 - 434
f) transmembrane 503 - 525
g) low complexity 569 - 582
h) transmembrane 616 - 638
i) transmembrane 643 - 662
j) transmembrane 674 - 696
k) transmembrane 716 - 734
l) transmembrane 758 - 775
m) transmembrane 807 - 829
(2) Transmembrane domains predicted by SOSUI: 11 transmembrane helices detected.
| No. | N terminal | transmembrane region | C terminal | type | length |
| 1 | 171 | VRWIKITGLFVFVVLCSILFSLY | 193 | PRIMARY | 23 |
| 2 | 335 | GVYTLIIFEIVHRTLAAMLGALA | 357 | PRIMARY | 23 |
| 3 | 379 | DFETLALLFGMMILVAVFSETGF | 401 | PRIMARY | 23 |
| 4 | 418 | WAMIFMLCLMAAILSAFLDNVTT | 440 | PRIMARY | 23 |
| 5 | 496 | MGLDFAGFTAHMFLGICLVLLVS | 518 | SECONDARY | 23 |
| 6 | 615 | RSLLVKCLTVLGFVISMFFLNSF | 637 | SECONDARY | 23 |
| 7 | 643 | LDLGWIAILGAIWLLILADIHDF | 665 | PRIMARY | 23 |
| 8 | 673 | EWATLLFFAALFVLMEALTHLHL | 695 | PRIMARY | 23 |
| 9 | 714 | QRFAAAIVLIVWVSALASSLIDN | 736 | PRIMARY | 23 |
| 10 | 763 | LMYALALGACLGGNGTLIGASTN | 785 | SECONDARY | 23 |
| 11 | 804 | FFRLGFPVMLMSCTIGMCYLLIA | 826 | PRIMARY | 23 |
(3) Graphical view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) Protein kinase C phosphorylation site : [occurs frequently]
38 - 40: TvK,
84 - 86: SlK,
89 - 91: SfK,
101 - 103: SqK,
149 - 151: SeK,
240 - 242: SgK,
448 - 450: TiR,
613 - 615: SdR.
b) ATP/GTP-binding site motif A (P-loop) : [occurs frequently]
58 - 65: AgeaaGKS
c) N-glycosylation site : [occurs frequently]
99 - 102: NSSQ,
210 - 213: NYSV,
214 - 217: NLSG,
269 - 272: NWTV,
437 - 440: NVTT,
749 - 752: NLSQ,
776 - 779: NGTL.
d) cAMP- and cGMP-dependent protein kinase phosphorylation site : [occurs frequently]
103 - 106: KKrS.
(2) Predicted results of subprograms by PSORT II:
a) Seems to have no N-terminal signal peptide
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: found, XXRR-like motif in the N-terminus: RLEN
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase: none.
(2) 3D models of mouse P protein predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=91,869Da, pI=6.21 (NP_068679).
FUNCTION
Ontology
a) Biological process: eye pigment biosynthesis
b) L-tyrosine, eye pigment precursor, and arsenite transporter activity
c) Integral to membrane
Location
Integral membrane protein.
Interaction
The P protein regulates posttranslational processing and transport of tyrosinase (Chen, et al, Toyofuku, et al).
No entries in the CuraGen Drosophila interaction database for CG15624.
Pathway
The P protein is believed to be an integral membrane protein involved in small molecule transport, specifically tyrosine - a precursor of melanin. P protein plays an important role in the generation or maintenance of melanosomal pH, which is required for the tyrosinase activity and the assembly of the normal melanogenic complex (Puri, et al). P protein increases cellular sensitivity to arsenicals and other metalloids and can modulate intracellular glutathione metabolism (Staleva, et al).
MUTATION
Allele or SNP
53 phenotypic alleles described in MGI:97454.
SNPs deposited in dbSNP.
Distribution
The p alleles are described in details in MGI:97454.
Effect
The transcript from this gene is missing or altered in six independent mutant alleles of the p locus (Gardner, et al)
PHENOTYPE
Mutations at the p gene cause the pink-eyed dilution (p), an autosomal recessive disorder similar to OCA2 (Gardner, et al; Rinchik, et al) (OMIM 203200). The p mutant shows a reduction of eumelanin (black-brown) pigment and altered morphology of black pigment granules (eumelanosomes), but have little effect on pheomelanin (yellow-red) pigment. For more details of the p phenotype, view the Mouse Locus Catalog #p. The original p mutation arose spontaneously. The pJ strain is described in more detail in JAX Mice database (C3H/HeJ-pJ/J).
REFERENCE
- Chen K, Manga P, Orlow SJ. Pink-eyed dilution protein controls the processing of tyrosinase. Mol Biol Cell 2002; 13: 1953-64. PMID: 12058062
- Gardner JM, Nakatsu Y, Gondo Y, Lee S, Lyon MF, King RA, Brilliant MH. The mouse pink-eyed dilution gene: association with human Prader-Willi and Angelman syndromes. Science 1992 ; 257: 1121-4. PMID: 1509264
- Puri N, Gardner JM, Brilliant MH. Aberrant pH of melanosomes in pink-eyed dilution (p) mutant melanocytes. J Invest Dermatol 2000; 115: 607-13. PMID: 10998131
- Rinchik EM, Bultman SJ, Horsthemke B, Lee ST, Strunk KM, Spritz RA, Avidano KM, Jong MT, Nicholls RD. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature 1993; 361: 72-6. PMID: 8421497
- Staleva L, Manga P, Orlow SJ. Pink-eyed dilution protein modulates arsenic sensitivity and intracellular glutathione metabolism. Mol Biol Cell 2002; 13: 4206-20. PMID: 12475946
- Toyofuku K, Valencia JC, Kushimoto T, Costin GE, Virador VM, Vieira WD, Ferrans VJ, Hearing VJ. The etiology of oculocutaneous albinism (OCA) type II: the pink protein modulates the processing and transport of tyrosinase. Pigment Cell Res 2002; 15: 217-24. PMID: 12028586