GENOMIC
Mapping
1qD, View the map and BAC clones (data from UCSC genome browser).
Structure
(assembly 10/03)
Mlph/NM_053015: 16 exons, 36,092bp, Chr1: 92,827,465-92,863,556.
Note that two shorter Mlph transcripts were also identified that seemed to result from the use of alternative poly(A)+ addition sites, which accounts for the three differently sized Mlph transcripts identified on Northern blots. All three Mlph transcripts encode identical proteins (Matesic, et al).
The figure below shows the structure of the Mlph gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Mlph (NM_053015), 4,229bp, view ORF and the alignment to genomic.
Note that two gaps in the genomic sequences exist in the last exon.
Expression Pattern
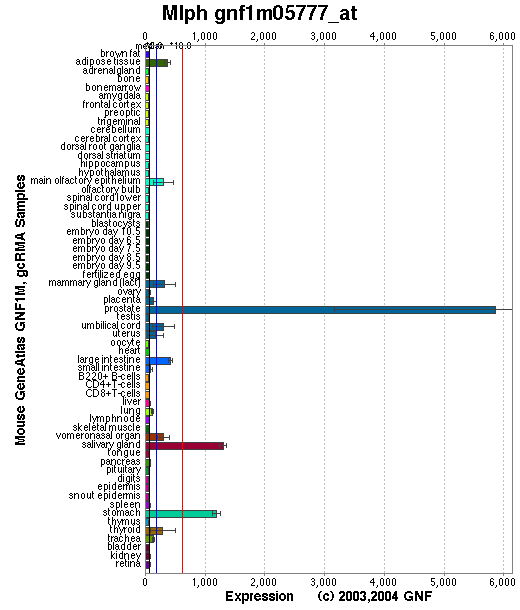
Tissue specificity: Highly expressed in embryos at day 7, not detectable at day 11. Highly expressed in adult stomach, detected at lower levels in kidney, lung, skin and small intestine. Detected in melanocytes. Not detectable in CTL cells (Hume, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
Melanophilin (NP_443748): 590aa, ExPaSy NiceProt view of Swiss-Prot:Q91V27.
Synonyms: Exophilin 3; Leaden protein; Synaptotagmin-like protein 2a; Slac2-a;
Slp homologue lacking C2 domains-a.
Ortholog
| Species | Human | Rat | Chicken | Fugufish |
| GeneView | GS3 | LOC316620 | 03904 | 156198 |
| Protein | NP_077006 (600aa) | XP_237388 (682aa) | 06194 (593aa) | 175372 (401aa) |
| Identities | 63% /386aa | 88% /482aa | 42% /262aa | 32% /66aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Motif/Site
(1) Predicted results by ScanProsite:
a) Rab-binding domain profile:
4 - 124: score=12.799
b) Amidation site : [occurs frequently]
1 - 4: mGKR,
571 - 574: nGRR.
c) N-glycosylation site : [occurs frequently]
60 - 63: NETH,
394 - 397: NISG.
d) cAMP- and cGMP-dependent protein kinase phosphorylation site : [occurs frequently]
345 - 348: KRmS,
495 - 498: RRkS,
541 - 544: RKyS,
573 - 576: RRgT.
e) Tyrosine sulfation site : [occurs frequently]
224 - 238:
qslsgepYsedttsl.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: Found XXRR-like motif in the N-terminus: GKRL
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase matched entries found, results here.
(2)ModBase Predicted Comparative 3D Structure of Q91V27 from UCSC Genome Sorter.
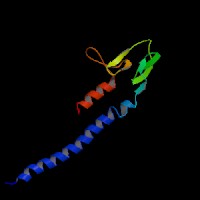
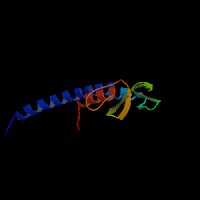
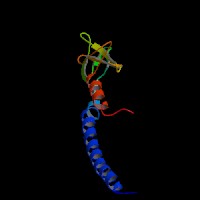
From left to right: Front, Top, and Side views of predicted protein
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=65,052Da, pI=5.70 (NP_443748).
FUNCTION
Ontology
a) Biological process: melanosome transport and melanocyte differentiation
b) Rab27A effector protein, actin binding, myosin binding
c) Cellular component: actin cytoskeleton
Location
Cytoplasmic. Associates with melanosomes.
Interaction
Melanophilin is one of the RAB27A effectors or a member of the exophilins or Slp/Slac2 family. It interacts with RAB27A via a conserved Rab27-binding domain, the Slp homology domain (SHD) (Strom, et al). The N-terminal SHD consists of two conserved alpha-helical regions (SHD1 and SHD2) that are often separated by two zinc finger motifs. SHD1 of melanophilin alone is both necessary and sufficient for high affinity specific recognition of the GTP-bound form of RAB27A. By contrast, the zinc finger motifs and SHD2 seem to be important for stabilization of the structure of the SHD or higher affinity RAB27A binding (Fukuda, et al (2002a); Nagashima, et al).
Slac2-a/melanophilin and Slac2-c/MyRIP are linker proteins between Rab27a and myosin Va. Slac2-a directly interacts with Rab27a and myosin Va via its N-terminal region (amino acids 1 to 146) and the middle region (amino acids 241 to 405), respectively (Fukuda, et al (2002b)). Melanophilin is required with Rab27a to recruit myosin Va to melanosomes in melanocytes (Hume, et al). Rab27a binds to the melanosome first and then recruits melanophilin, which in turn recruits myosin-Va. Melanophilin creates this link by binding to Rab27a in a GTP-dependent fashion through its amino terminus, and to myosin-Va through its carboxy terminus (Wu, et al). GTP-hydrolysis leads to the inactivation of Rab27a and presumably to the dissociation of melanophilin and myosin Va. cAMP stimulates the expression of Rab27a and rapidly increases the interaction of the melanophilin/Slac2-a with actin, allowing the rapid accumulation of melanosomes in the actin-rich region of the dendrite extremities after the action of melanocyte-differentiating agent such as alpha-melanocyte-stimulating hormone (Passeron, et al). Melanophilin directly activates the actin-activated ATPase activity of myosin Va and thus its motor activity (Li, et al).
The most C-terminal conserved region of Slac2-a (amino acids 400-590) and Slac2-c (amino acids 670-856), which is not essential for myosin Va binding, directly binds actin. Expression of these regions in PC12 cells and melanoma cells colocalized with actin filaments at the cell periphery, suggesting a novel role of Slac2-a/c in capture of Rab27-containing organelles in the actin-enriched cell periphery (Fukuda, et al (2002c)).
No MLPH drosophila homolog shown in CuraGen interaction database.
Pathway
Involved in melanosome transport and distribution (view diagram of the Slac2-a tripartite protein complex and the dynamics of melanosomes here).
MUTATION
Allele or SNP
2 phenotypic alleles described in MGI: 2176380.
SNPs deposited in dbSNP.
Distribution
| Location | Genomic | cDNA | Protein | Type | Strain | Reference |
| Exon 2 | 91C>T | 90G~110A del; 91C>T | R31del 21bp (7aa); R31X | splicing + nonsense | ln (C57L/J) | Matesic, et al |
Effect
The 91C>T mutation in ln is predicted as a null mutation. The Mlph coding region is completely deleted in lnllRK3 DNA and Mlph transcripts are not expressed from the lnllRK3 chromosome (Matesic, et al).
PHENOTYPE
The Mlph mutations cause an autosomal recessive disorder, leaden (ln) (Matesic, et al), resembling Griscelli syndrome type 3 (GS3) (Menasche, et al) (OMIM:606526). Ln arose spontaneously in the C57BR inbred strain. The strain is described in more detail in JAX Mice database (C57L/J-Mlphln). The effect on coat color of ln/ln is indistinguishable from the dilute (d/d) mouse. Like dilute it causes clumping of melanin granules into larger masses, but no change in color of the pigment. The clumping is due to the shape of the melanocytes, which have fewer and thinner dendritic processes than wild-type melanocytes. These melanocytes are more easily dislodged from fixed sites in the hair bulb and incorporated into the developing hair, resulting in large clumps of pigment in the hair shaft (Mouse Locus Catalog #Mlph).
The Mlphln-l1Rk3 allele was induced by triethylene melamine. Homozygotes of Mlphln-l1Rk3 are lethal. When balanced over ln (ln-llRK3/ln), it produces a phenotype that is similar to homozygous ln mice.
REFERENCE
- Fukuda M. Synaptotagmin-like protein (Slp) homology domain 1 of Slac2-a/melanophilin is a critical determinant of GTP-dependent specific binding to Rab27A. J Biol Chem 2002a; 277: 40118-24. PMID: 12189142
- Fukuda M, Kuroda TS, Mikoshiba K. Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: implications of a tripartite protein complex for melanosome transport. J Biol Chem 2002b; 277: 12432-6. PMID: 11856727
- Fukuda M, Kuroda TS. Slac2-c (synaptotagmin-like protein homologue lacking C2 domains-c), a novel linker protein that interacts with Rab27, myosin Va/VIIa, and actin. J Biol Chem 2002c; 277: 43096-103. PMID: 12221080
- Hume AN, Collinson LM, Hopkins CR, Strom M, Barral DC, Bossi G, Griffiths GM, Seabra MC. The leaden gene product is required with Rab27a to recruit myosin Va to melanosomes in melanocytes. Traffic 2002; 3: 193-202. PMID: 11886590
- Li XD, Ikebe R, Ikebe M. Activation of Myosin Va function by melanophilin, a specific docking partner of Myosin Va. J Biol Chem 2005; 280:17815-22. PMID: 15760894
- Matesic LE, Yip R, Reuss AE, Swing DA, O'Sullivan TN, Fletcher CF, Copeland NG, Jenkins NA. Mutations in Mlph, encoding a member of the Rab effector family, cause the melanosome transport defects observed in leaden mice. Proc Natl Acad Sci U S A 2001; 98: 10238-43. PMID: 11504925
- Menasche G, Ho CH, Sanal O, Feldmann J, Tezcan I, Ersoy F, Houdusse A, Fischer A, de Saint Basile G. Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5A F-exon deletion (GS1). J Clin Invest 2003; 112: 450-6. PMID: 12897212
- Nagashima K, Torii S, Yi Z, Igarashi M, Okamoto K, Takeuchi T, Izumi T. Melanophilin directly links Rab27a and myosin Va through its distinct coiled-coil regions. FEBS Lett 2002; 517: 233-8. PMID: 12062444
- Passeron T, Bahadoran P, Bertolotto C, Chiaverini C, Busca R, Valony G, Bille K, Ortonne JP, Ballotti R. Cyclic AMP promotes a peripheral distribution of melanosomes and stimulates melanophilin/Slac2-a and actin association. FASEB J 2004; 18: 989-91. PMID: 15059972
- Strom M, Hume AN, Tarafder AK, Barkagianni E, Seabra MC. A family of Rab27-binding proteins. Melanophilin links Rab27a and myosin Va function in melanosome transport.J Biol Chem 2002; 277: 25423-30. PMID: 11980908
- Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenkins NA, Hammer JA 3rd. Identification of an organelle receptor for myosin-Va. Nat Cell Biol 2002; 4: 271-8. PMID: 11887186
EDIT HISTORY:
Created by Wei Li & Jonathan Bourne: 07/20/2004
Updated by Wei Li: 04/06/2006