GENOMIC
Mapping
5qF. View the map and BAC contig (data from UCSC genome browser).
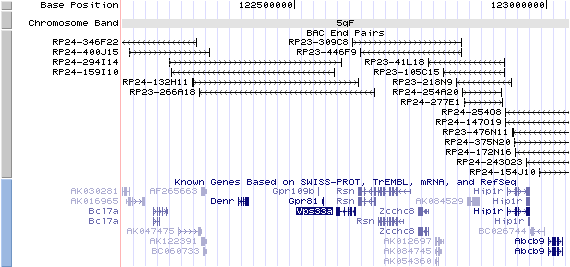
Structure
(assembly 10/03)
Vps33a (NM_029929): 13 exons, 38,960bp, chr5:122,580,968-122,619,927.
The figure below shows the structure of the Vps33a gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Vps33a/NM_029929: 3,853bp, view ORF and the alignment to genomic.
Expression Pattern
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.

PROTEIN
Sequence
Vacuolar protein sorting 33A (NP_084205): 598aa, ExPaSy NiceProt view of Swiss-Prot:Q9D2N9.
Synonym: Vps33p.
Ortholog
| Species | Human | Rat | Zebrafish | Fruitfly | Yeast |
| GeneView | VPS33A | Vps33a | 10902 | car | VPS33 |
| Protein | NP_075067 (596aa) | NP_075250 (597aa) | 13152 (577aa) | AAD38513 (617aa) | Vps33p (691aa) |
| Identities | 97%/582aa | 97%/586aa | 76%/459aa | 43%/265aa | 27%/117aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. More homologs shown in HomoloGen:11294.
Domain
(1) Domains predicted by SMART:
a) low complexity: 12 - 27
b) low complexity: 225 - 237
(2) Transmembrane domains predicted by SOSUI: none.
(3) CDD domain: KOG1302: Vacuolar sorting protein VPS33/slp1 (Sec1 family) [Intracellular trafficking, secretion, and vesicular transport].
(4) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site [pattern] [Warning: pattern with a high probability of occurrence]:
355 - 358 NHTS.
b) Tyrosine sulfation site [rule] [Warning: rule with a high probability of occurrence]:
157 - 171 egafkecYlegdqts, 291 - 305 lnsaeelYaeirdkn.
c) Protein kinase C phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
65 - 67 TlK, 314 - 316 SkK, 388 - 390 TdK, 467 - 469 TiR, 471 - 473 TlR, 498 - 500 SvR, 580 - 582 TtK.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: KKXX-like motif in the C-terminus: MEKP
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: too long tail
i) Dileucine motif in the tail: found - LL at 54.
3D Model
(1) ModBase: predicted comparative 3D structures on Q9D2N9 (data from UCSC Gene Sorter). (from left to right: Front, Top, Side view)
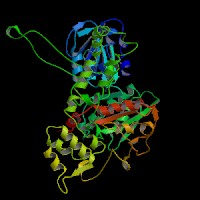
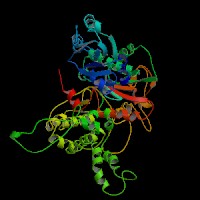
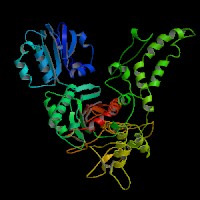
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
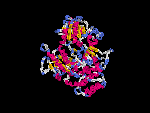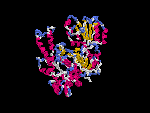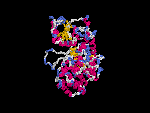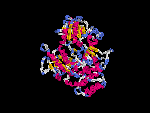
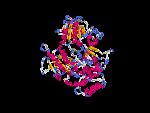
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=67,527Da, pI=6.64.
FUNCTION
Ontology
a) Process: Golgi-to-vacuole protein transport, protein secretion (overview of trafficking pathway here).
b) Belongs to the STXBP/UNC-18/SEC1 family.
c) Component of membrane.
Location
Cytoplasmic, peripheral membrane protein associated with late endosomes/lysosomes. Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner (Gerhardts, et al; Banta, et al).
Interaction
This gene is a member of the Sec-1 domain family, and it encodes a protein similar to the yeast class C Vps33 protein. A large hetero-oligomeric class C Vps complex consists of VPS33 together with VPS11, VPS16, VPS18, and probably also the VPS39 and VPS41 proteins (Huizing, et al; Rieder, et al ). In yeast, a 38S complex consisting of both Vam2/6p and the class C Vps proteins, which is termed as HOPS, is responsible for homotypic fusion and vacuole protein sorting (Seals, et al). The C-Vps complex binds to Vam3-Vti1-Vam7 paired SNARE complexes and STX7 (Sato, et al). At the endosome, Vps33p and other class C members exist as a complex with Vps8p, a protein previously known to act in transport between the late Golgi and the endosome. C. elegans SPE-39 interacts in vitro with both VPS33A and VPS33B, whereas RNA interference of VPS33B causes spe-39-like spermatogenesis defects. The human SPE-39 orthologue C14orf133 also interacts with VPS33 homologues and both coimmunoprecipitates and cosediments with other HOPS subunits. SPE-39 knockdown in cultured human cells altered the morphology of syntaxin 7-, syntaxin 8-, and syntaxin 13-positive endosomes (Zhu, et al).
Vps33p also interacts with Pep12p, a known interactor of the SM protein Vps45p (Subramanian, et al). A novel endosomal tethering (CORVET) complex, which interacts with the Rab GTPase Vps21, has been described. Both HOPS and CORVET complexes share the four class C Vps proteins: Vps11, Vps16, Vps18, and Vps33. The HOPS complex, in addition, contains Vps41/Vam2 and Vam6, whereas the CORVET complex has the Vps41 homolog Vps8 and the (h)Vam6 homolog Vps3. The CORVET and HOPS complexes can interconvert which leads to two additional intermediate complexes, both consisting of the class C core bound to Vam6-Vps8 or Vps3-Vps41 (Peplowska, et al). In drosophila, Deep orange and carnation reside in a protein complex that localizes to endosomal compartments (Sevrioukov, et al).
Vps33a interacts with the cytoplasmic tail of RANKL (Kariya, et al). Drosophila vps33a/CAR interacts with syntaxin 16 (Akbar, et al).
7 proteins are shown to be associated with YLR396C/VPS33 in Yeast GRID.
No entry for the Vps33a drosophila homolog CG12230/car in CuraGen interaction database.
Pathway
The Class C Vps complex plays essential roles in the processes of membrane docking and fusion at both the Golgi-to-endosome and endosome-to-vacuole stages of transport (view diagram of Class C Vps pathway here). (Peterson, et al; Sato, et al; Pevsner, et al ). Vps33p functions at multiple trafficking steps and is not limited to action at the vacuolar membrane (Subramanian, et al).
MUTATION
Allele or SNP
1 phenotypic allele is described in MGI:1924823.
SNPs deposited in dbSNP.
Distribution
| Location | Genomic | cDNA | Protein | Type | Strain | Reference |
| Exon 6 | 753T>G | 753T>G | D251E | missense | bf (B6) | Suzuki, et al |
Effect
D251 is highly conserved (refer to the multiple sequence alignment PDF file). This residule is located only three residues distal to the Gly disrupted by the carnation (car) mutation (G249V) in Drosophila. Suzuki, et al further confirmed that the phenotype caused by the D251E mutation was completely complemented by wild-type expression in stably transfected melanocytes.
PHENOTYPE
Mutation in the Vps33a gene is the cause of buff mutant (Suzuki, et al), a mouse model of Hermansky-Pudlak syndrome (OMIM 203300). The bf allele arose from C57BL/6J. On an agouti background, homozygotes have khaki colored coats and show higher than normal levels of lysosomal glycosidases in their kidneys. Melanosomes of the RPE and choroid are reduced in both number and size in the Vps33a/bf mutant (Suzuki, et al). Buff mice showed a novel phenotype observed in urothelial umbrella cells, where the uroplakin-delivering FVs were almost completely replaced by Rab27b-negative multivesicular bodies (MVBs) involved in uroplakin degradation(Guo, et al). In addition, mutation of Vps33a affects the cell surface expression level of RANKL and disrupts the trafficking of RANKL to the secretory lysosomes in osteoblasts or bone marrow stromal cells (BMSCs) (Kariya, et al).
The bf mice demonstrated significant motor deficits (e.g., grip strength, righting reflex and touch escape) worsening with age, progressive Purkinje cell loss, and reduced (~30%) cerebella size (Chintala, et al). The mutant is described in more detail in JAX Mice database (C57BL/6J-Vps33abf/J) and Mouse Locus Card #Vps33a.
REFERENCE
- Akbar MA, Ray S, Kr?mer H. The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol Biol Cell 2009; 20: 1705-14. PMID: 19158398
- Banta LM, Vida TA, Herman PK, Emr SD. Characterization of yeast Vps33p, a protein required for vacuolar protein sorting and vacuole biogenesis. Mol Cell Biol 1990; 10: 4638-49. PMID: 2201898
- Chintala S, Novak EK, Spernyak JA, Mazurchuk R, Torres G, Patel S, Busch K, Meeder BA, Horowitz JM, Vaughan MM, Swank RT. The Vps33a gene regulates behavior and cerebellar Purkinje cell number. Brain Res 2009; 1266:18-28.PMID: 19254700
- Gerhardt B, Kordas TJ, Thompson CM, Patel P, Vida T. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J Biol Chem 1998; 273: 15818-29. PMID: 9624182
- Guo X, Tu L, Gumper I, Plesken H, Novak EK, Chintala S, Swank RT, Pastores G, Torres P, Izumi T, Sun TT, Sabatini DD, Kreibich G. Involvement of vps33a in the fusion of uroplakin-degrading multivesicular bodies with lysosomes. Traffic 2009; 10: 1350-61. PMID: 19566896
- Kariya Y, Honma M, Aoki S, Chiba A, Suzuki H. Vps33a mediates RANKL storage in secretory lysosomes in osteoblastic cells. J Bone Miner Res 2009; 24: 1741-52. PMID: 19419298
- Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell 2007; 12:739-50. PMID: 17488625
- Peterson MR, Emr SD. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic 2001; 2: 476-86. PMID: 11422941
- Pevsner J, Hsu SC, Hyde PS, Scheller RH. Mammalian homologues of yeast vacuolar protein sorting (vps) genes implicated in Golgi-to-lysosome trafficking. Gene 1996; 183: 7-14. PMID: 8996080
- Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell 1997; 8: 2307-27. PMID: 9362071
- Sato TK, Rehling P, Peterson MR, Emr SD. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol Cell 2000; 6: 661-71. PMID: 11030345
- Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci U S A 2000; 97: 9402-7. PMID: 10944212
- Sevrioukov EA, He JP, Moghrabi N, Sunio A, Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell 1999; 4: 479-86. PMID: 10549280
- Subramanian S, Woolford CA, Jones EW. The Sec1/Munc18 protein, Vps33p, functions at the endosome and the vacuole of Saccharomyces cerevisiae. Mol Biol Cell 2004; 15: 2593-605. PMID: 15047864
- Suzuki T, Oiso N, Gautam R, Novak EK, Panthier JJ, Suprabha PG, Vida T, Swank RT, Spritz RA. The mouse organellar biogenesis mutant buff results from a mutation in Vps33a, a homologue of yeast vps33 and Drosophila carnation. Proc Natl Acad Sci U S A 2003; 100: 1146-50. PMID: 12538872
- Zhu GD, Salazar G, Zlatic SA, Fiza B, Doucette MM, Heilman CJ, Levey AI, Faundez V, L'hernault SW. SPE-39 family proteins interact with the HOPS complex and function in lysosomal delivery. Mol Biol Cell 2009; 20: 1223-40. PMID: 19109425
EDIT HISTORY:
Created by Wei Li & Jonathan Bourne 06/28/2004
Updated by Wei Li, 02/28/2008
Updated by Wei Li, 03/12/2009
Updated by Wei Li, 05/25/2011