GENOMIC
Mapping
4qC3 View the map and BAC clones (data from UCSC genome browser).
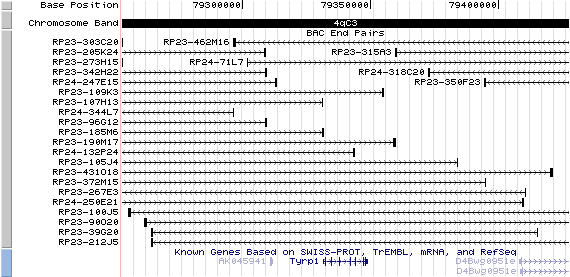
Structure
(assembly 10/03)
Tyrp1/NM_031202: 8 exons, 17,490bp, Chr4: 79,331,424 - 79,348,910.
The figure below shows the structure of the Tyrp1 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
The flanking DNA of TRP-1, unlike tyrosinase, does not contain a TATA box or a CCAAT box. Both mouse genes, however, share an 11bp sequence, also found in human tyrosinase, which may be a melanocyte-specific promoter element (Jackson, et al).
TRANSCRIPT
RefSeq/ORF
Tyrp1 (NM_031202), 2,740bp, view ORF and the alignment to genomic.
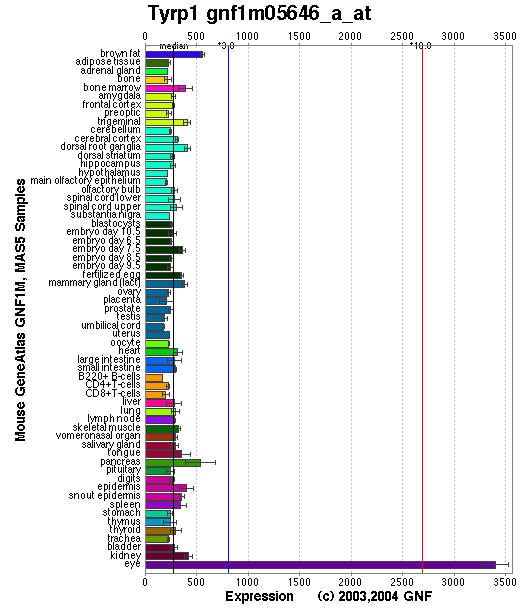
Expression Pattern
Tissue specificity: Pigment cells.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
Tyrosinase-related protein 1 (NP_112479): 537aa, ExPaSy NiceProt view of Swiss-Prot:P07147.
Synonym: Brown locus protein; 5,6-dihydroxyindole-2-carboxylic acid oxidase [Precursor] (DHICA oxidase); TRP-1; TRP1; TRP; Catalase B; Glycoprotein-75 (gp75); Melanoma antigen gp75.
Ortholog
| Species | Human | Cow | Rat | Chicken |
| GeneView | OCA3 | TYRP1 | Tyrp1 | LOC395913 |
| Protein | NP_000541 (537aa) | NP_776905 (537aa) | XP_238398 (537aa) | NP_990376 (535aa) |
| Identities | 85% /458aa | 84% /456aa | 93% /501aa | 73% /390aa |
View multiple sequence alignment (PDF files) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) signal peptide 1-25
b) Pfam: Tyrosinase 121-471
c) tansmembrane 479-501
(2) Transmembrane domains predicted by SOSUI: Two transmembrane helices detected.
| No. | N terminal | transmembrane region | C terminal | type | length |
| 1 | 1 | MFLAVLYCLLWSFQISDGHFPR | 22 | SECONDARY | 22 |
| 2 | 476 | PWLLGAALVGAVIAAALSGLSSR | 498 | PRIMARY | 23 |
(3) Graphical view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) EGF-like domain signature 1 :
99 - 110: CqCndnfsGhnC
b) Laminin-type EGF-like (LE) domain signature :
99 - 122: Cq.CndnfsGhnCgtCrpg.Wrgaa...........C
c) Tyrosinase CuA-binding region signature :
215 - 232: Hegpa.FLtWHRyhLlqlE
d) Tyrosinase and hemocyanins CuB-binding region signature :
397 - 408: DPiFVllHtftD
e) N-myristoylation site : [occurs frequently]
52 - 57: GTdpCG,
57 - 62: GSssGR,
119 - 124: GAacNQ,
267 - 272: GSrsNF,
300 - 305: GTlcNS,
470 - 475: GQefTV,
495 - 500: GVasCL.
f) Protein kinase C phosphorylation site : [occurs frequently]
60 - 62: SgR,
112 - 114: TcR,
128 - 130: TvR,
163 - 165: TrR,
195 - 197: SvK,
253 - 255: TgK,
354 - 356: SfR,
366 - 368: TgK,
505 - 507: StK.
g) N-glycosylation site : [occurs frequently]
96 - 99: NRTC,
104 - 107: NFSG,
181 - 184: NISV,
304 - 307: NSTE,
350 - 353: NSTD,
385 - 388: NGTG,
533 - 536: NHSM.
h) cAMP- and cGMP-dependent protein kinase phosphorylation site : [occurs frequently]
152 - 155: KRtT
i) Tyrosine sulfation site : [occurs frequently]
289 - 303:
vcesleeYdtlgtlc,
519 - 533:
yqryaedYeelpnpn
(2) Predicted results of subprograms by PSORT II:
a) Seems to have a cleavable signal peptide (1 to 24)
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: found at 518, 521, 525
i) Dileucine motif in the tail: found LL at 514
3D Model
(1) ModBase entry found, results here.
(2)ModBase Predicted Comparative 3D Structure of P07147 from UCSC Genome Sorter.
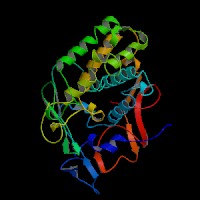
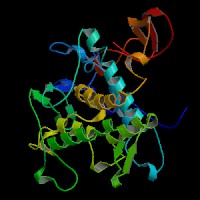
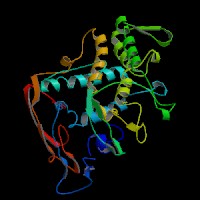
From left to right: Front, Top, and Side views of predicted protein
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=60,761Da, pI=5.65 (NP_112479).
FUNCTION
Ontology
a) Biological process: melanin biosynthesis from tyrosine
b) Monooxygenase activity; oxidoreductase activity
c) Integral to membrane
d) Belongs to the tyrosinase family
Location
Type I membrane protein. Melanosome.
Interaction
Tyrosinase-related protein 1 (Tyrp1) is involved in maintaining stability of tyrosinase protein and modulating its catalytic activity. TYRP1 binds 2 copper ions per subunit via CuA and CuB motifs. Phosphorylation of tyrosinase by PKC-beta induces a complex formation between tyrosinase and TYRP1 (Wu, et al).
Pathway
Tyrp1 is a melanocyte-specific gene product involved in eumelanin synthesis. It functions in the oxidation of 5,6-dihydroxyindole-2-carboxylic acid (DHICA) into indole-5,6-quinone-2-carboxylic acid (Olivares, et al). Tyrp1 may play a significant role in mediating ethnic differences in melanogenesis and constitutive skin pigmentation in vivo (Alaluf, et al). Tyrp1 plays a role in the distal pigment eumelanin pathway. Tyrp1 is sorted to the melanosomes for melanin synthesis with an AP-3 complex independent pathway (Huizing, et al) (view diagram of melanosome maturation in melanocytes here).
KEGG Biochemical Pathways:
hsa00350 - Tyrosine metabolism - Homo sapiens
MUTATION
Allele or SNP
49 phenotypic alleles described in MGI:98881.
SNPs deposited in dbSNP.
Distribution
Among the 9 spontaneous mutations, the allelic b and B-lt have been characterized molecularly (see details in MGI:98881).
Effect
The identified 2 spontaneous mutations so far are missense mutations which may disrupt the function of tyrosinase-related protein 1.
PHENOTYPE
Defects in Tyrp1 are the cause of the brown (b) mutant, an autosomal recessive disorder similar to OCA3 (OMIM 203290). Brown mice have a brown or hypopigmented coat. Loss of the Tyrp1 leads to changes of the amount and type of eumelanin, but does not result in the complete loss of eumelanin or pheomelanin. Homozygous recessive mutants on a nonagouti/black background are chocolate brown and show altered melanin granule morphology. Semidominant mutants result in melanocyte degeneration causing reduced pigmentation and progressive hearing loss. For more details of the brown phenotype, view the Mouse Locus Catalog #Tyrp1. The b mutation arose spontaneously from C57BL. The strain is described in more detail in JAX Mice database (C57BL/6J-Tyrp1b-J/J).
In brown dogs, mutations in the canine TRYP1 gene interfere with TYRP1 function and cause the diluted coat color (Schmutz, et al).
REFERENCE
- Alaluf S, Barrett K, Blount M, Carter N. Ethnic variation in tyrosinase and TYRP1 expression in photoexposed and photoprotected human skin. Pigment Cell Res 2003; 16: 35-42. PMID: 12519123
- Huizing M, Sarangarajan R, Strovel E, Zhao Y, Gahl WA, Boissy RE. AP-3 mediates tyrosinase but not TRP-1 trafficking in human melanocytes. Mol Biol Cell 2001; 12: 2075-85. PMID: 11452004
- Jackson IJ, Chambers DM, Budd PS, Johnson R. The tyrosinase-related protein-1 gene has a structure and promoter sequence very different from tyrosinase. Nucleic Acids Res 1991; 19: 3799-804. PMID: 1907365
- Olivares C, Jimenez-Cervantes C, Lozano JA, Solano F, Garcia-Borron JC. The 5,6-dihydroxyindole-2-carboxylic acid (DHICA) oxidase activity of human tyrosinase. Biochem J 2001; 354: 131-9. PMID: 11171088
- Schmutz SM, Berryere TG, Goldfinch AD. TYRP1 and MC1R genotypes and their effects on coat color in dogs. Mamm Genome 2002; 13: 380-7. PMID: 12140685
- Wu H, Park HY. Protein kinase C-beta-mediated complex formation between tyrosinase and TRP-1. Biochem Biophys Res Commun 2003; 311: 948-53. PMID: 14623273