GENOMIC
Mapping
11q14.3. View the map and BAC clones (data from UCSC genome browser).
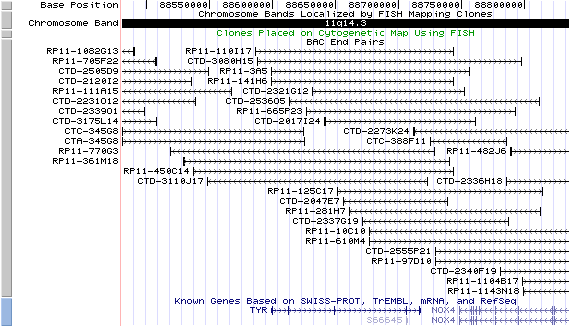
Structure
(assembly 07/03)
TYR/NM_000372: 5 exons, 118,207bp, Chr11: 88,598,773 - 88,716,979.
The figure below shows the structure of the TRY gene (data from UCSC genome browser).
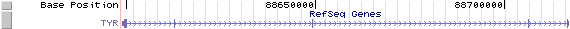
Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
TYR (NM_000372), 2,384bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: Highest expression in skin tissue. Increased expression after UV-B radiation.
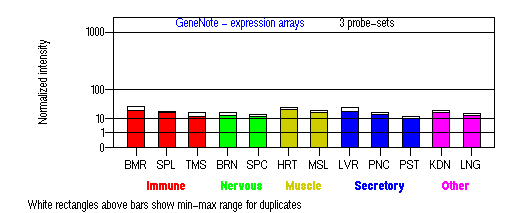
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle;
LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
Tyrosinase (NP_000363): 529aa, ExPaSy NiceProt view of Swiss-Prot:P14679.
Synonyms: albino protein homolog.
Enzyme: EC 1.14.18.1.
Ortholog
| Species | Mouse | Cow | Rat |
| GeneView | Tyr/c | TYR | Tyr |
| Protein | NP_035791 (533aa) | NP_851344(530aa) | XP_238901(530aa) |
| Identities | 85% /457aa | 86% /461aa | 87% /462aa |
| Species | Chicken | Zebrafish | Worm |
| GeneView | TYR | tyr | C34G6.2 |
| Protein | NP_989491(529aa) | NP_571088(535aa) | WP:CE08574(751aa) |
| Identities | 73% /389aa | 61% /324aa | 27% /73aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) signal peptide 1-18
b) Pfam: Tyrosinase 111-456
c) tansmembrane 474-498
(2) Transmembrane domains predicted by SOSUI: Two transmembrane helices detected.
| No. | N terminal | transmembrane region | C terminal | type | length |
| 1 | 1 | MLLAVLYCLLWSFQTSAGHFPR | 22 | PRIMARY | 22 |
| 2 | 476 | SWLLGAAMVGAVLTALLAGLVSL | 498 | PRIMARY | 23 |
(3) Graphical view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) EGF-like domain signature 1 :
89 - 100: CqCsgnfmGfnC
b) Laminin-type EGF-like (LE) domain signature :
89 - 112: Cq.CsgnfmGfnCgnCkfgfWgpn............C
c) Tyrosinase CuA-binding region signature :
202 - 219: Heapa.FLpWHRlfLlrwE
d) Tyrosinase and hemocyanins CuB-binding region signature :
383 - 394: DPiFLlhHafvD
e) Protein kinase C phosphorylation site : [occurs frequently]
26 - 28: SsK,
50 - 52: SgR,
113 - 115: TeR,
309 - 311: TpR,
339 - 341: SfR,
441 - 443: SsK.
f) N-glycosylation site : [occurs frequently]
86 - 89: NRTC,
111 - 114: NCTE,
161 - 164: NGST,
230 - 233: NFTI,
337 - 340: NFSF,
371 - 374: NGTM.
g) Microbodies C-terminal targeting signal : [occurs frequently]
527 - 529: SHL
h) Tyrosine sulfation site : [occurs frequently]
442 - 456:
skdlgydYsylqdsd
Note: The above scan shows two binding regions which correspond to the 2 copper ion cofactors present per subunit
(2) Predicted results of subprograms by PSORT II:
a) Seems to have a cleavable signal peptide (1 to 17)
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: found at 520 and 524
i) Dileucine motif in the tail: found LL at 514
3D Model
(1) ModBase entry found, results here.
(2)ModBase Predicted Comparative 3D Structure of P14679 from UCSC Genome Sorter.
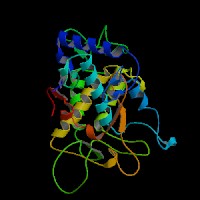
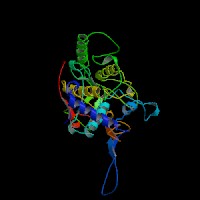

From left to right: Front, Top, and Side views of predicted protein
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=60,393Da, pI=5.71 (NP_000363, precursor).
FUNCTION
Ontology
a) Biological process: eye pigment biosynthesis; melanin biosynthesis from tyrosine
b) Biological process: visual perception
c) Monophenol monooxygenase activity
d) Component of Golgi vesicle
e) Integral to membrane
Location
Type I membrane protein. Melanosome.
Interaction
Tyrosinase binds 2 copper ions per subunit (CuA and CuB). Tyrosinase is activated when protein kinase C-beta (PKC-beta) phosphorylates the serine residues at amino acid positions 505 and 509. Phosphorylation of tyrosinase by PKC-beta induces a complex formation between tyrosinase and TYRP1 (Wu, et al). The p protein regulates posttranslational processing of tyrosinase, and hypopigmentation in melan-p1 cells is the result of altered tyrosinase processing and trafficking (Chen, et al, Toyofuku, et al).
Pathway
Tyrosinase (Monophenol monooxygenase, EC 1.14.18.1) is a glycoprotein and a copper-containing oxidase that functions in the formation of pigments such as melanins and other polyphenolic compounds. It catalyzes the rate-limiting conversions of tyrosine to dopa, dopa to dopa-quinone and possibly 5,6-dihydroxyindole to indole-5,6 quinone (L-tyrosine + L-DOPA + O2 = L-DOPA + DOPAquinone + H2O).
During the melanogenesis, tyrosinase can be released from the ER in the presence of protonophore or proton pump inhibitors which increase the pH of intracellular organelles, after which tyrosinase is transported correctly to the Golgi, and then to melanosomes via the endosomal sorting system (Watabe, et al). Tyrosinase is sorted via the dilucine signal to the melanosomes for melanin synthesis mediated by AP-3 complex (view diagram of melanosome maturation in melanocytes here).
KEGG Biochemical Pathways:
(1) hsa00350 - Tyrosine metabolism - Homo sapiens
(2) hsa00940 - Flavonoids, stilbene and lignin biosynthesis - Homo sapiens
MUTATION
Allele or SNP
219 mutations deposited in HGMD.
SNPs deposited in dbSNP.
Selected allelic examples described in OMIM.
Distribution
The distribution of mutations and polymorphisms is described in detail in Albinism Database. For cross-reference, view the distribution of mutations and polymorphisms in the Retina International Mutation Database of TYR Gene.
54 novel mutations not appeared in the HGMD database:
(1) Chaki, et al (2006) reported 5 novel mutations (D42N, C91S, E219K, Q326X, G372R) in Indian OCA1 patients.
(2) Hutton, et al (2008a) analyzed Caucasian patients with autosomal recessive ocular albinism (AROA). All were compound heterozygotes with one R402Q allele. One novel mutation (c.47delC) was reported.
(3) Hutton, et al (2008b) reported 8 novel mutations, one frameshift (c.124delG) and seven missense substitutions (F84V, I123T, Y149C, Y181C, H202R, P209L, and L288F) in Caucasian OCA1 patients. They excluded the P152S from a pathological mutation. Among the non-Hispanic Caucasian OCA1 patients, the T373K mutation was most frequent (13.7%). T373K is by far the most frequent OCA1A mutant allele, followed by P81L. V275F and IVS2-7T are the most frequent OCA1B mutant alleles.
(4) Wang, et al (2009) reported 4 novel mutations (C24Y, K142M, G315X, W475X) in Chinese OCA1 patients and first reported two known mutations (
c.896G>A and c.230G>A) in Chinese OCA1 patients.
(5) Rooryck, et al (2008) reported 13 novel ones: three splice site mutations (c.1185 -1 G>T, c.1366 +1 G>A, c.1366 +3A>T), three nonsense mutation (S163X, E219X, K465X), three missense mutations (H19R, K33T, W400C), one frameshift (c.820 -1_820GA>TG, one deletion of three nucleotides (c.505_507delCAG), one deletion of the entire gene (11q14.3 deletion), and one deletion of a single exon (Exon 2 deletion).
(6) Gronskov, et al (2009) reported 8 novel mutations (D76E, A206T, Q255X, C276Y, R299C, M332I, V427F, L452V) in Danish OCA or AROA patients.
(7) Wei, et al (2010) reported 15 previously unreported alleles:
p.C24R, p.W39C, p.W39R, p.R77G, p.C100F, p.C100W, p.M185V, p.H211R, p.R212T, p.Y235H, p.W236R, p.D249G, c.1146C>G (p.N382K), p.A481E, and p.G506X in Chinese OCA patients. Interestingly, c.575C>A (p.S192Y) has been reported as a single-nucleotide polymorphism (SNP),
rs1042602, in several populations. However, the A allele was not found in any of the 100 unaffected subjects from the Chinese Han population. Two alleles, p.R299H and c.929insC, are the most common alleles in Chinese OCA1 patients, accounting for 15.4% and 14.8%,
respectively. About 81% alleles were clustered on exons 1 and 2 of TYR gene in this population.
Note that the 3' region (about 68 kb) of TYR, encompassing exons 4 and 5, has 98.55% sequence identity with a pseudogene (TYRL, 11p11.2, OMIM 191270). A PCR strategy to amplify TYR and TYRL separately was developed by Chaki et al. (2005). They found that the previously reported mutational allele c.1217C4T (p.P406L) was identified as an allele from the pseudogene, TYRL.
Effect
The majority of TYR mutations are missense or nonsense mutations. A large number of mutations have been identified in tyrosinase, with many leading to its misfolding, endoplasmic reticulum (ER) retention, and degradation. Many of the missense mutations are clustered in or flank the copper binding sites by affecting copper binding or by disrupting the substrate binding site (Spritz, et al). The G97R missense mutation is the first identified within the epidermal growth factor (EGF)-like sequence and the H19Q missense mutation alters the cleavage site of the signal peptide sequence (Oetting, et al).
PHENOTYPE
Defects in TYR (OMIM 606933) are the cause of oculocutaneous albinism type IA (OCA-IA) (OMIM 203100), also known as oculocutaneous albinism type I (OCA-I). OCA-IA patients have no melanin synthesis with white skin and hair. In oculocutaneous albinism, the reduction in melanin pigment in the skin results in an increased sensitivity to ultraviolet radiation and to predisposition to skin cancer.
Defects in TYR are the cause of oculocutaneous albinism type IB (OCA-IB) (OMIM 606952), also known as albinism yellow mutant type. OCA-IB patients have white hair at birth that rapidly turns yellow or blond.
Defects in TYR are the cause of oculocutaneous albinism type I temperature sensitive (OCA-ITS) (OMIM 606952). OCA-ITS patients have white axilary and scalp hair and pigmented arm and leg hair.
Individuals with OCA1 show a complete absence of pigment in their skin, eyes and hair; this persists throughout life in the subtype OCA1A, whereas patients with subtype OCA1B have some residual tyrosinase activity and thus gradually accumulate minor amounts of pigment in their tissues.
Compound heterozygosity for the R402Q polymorphism and a mutant allele of TYR is a common cause of autosomal recessive ocular albinism. The R402Q polymorphism is also found in Waardenburg syndrome type II with ocular albinism (WS2-OA, OMIM 103470) in association with a deletion in the MITF gene.
REFERENCE
- Chaki M, Mukhopadhyay A, Ray K. Determination of variants in the 3'-region of the tyrosinase gene requires locus specific amplification. Hum Mutat 2005; 26:53-8.PMID: 15895460
- Chaki M, Sengupta M, Mukhopadhyay A, Subba Rao I, Majumder PP, Das M, Samanta S, Ray K. OCA1 in different ethnic groups of india is primarily due to founder mutations in the tyrosinase gene. Ann Hum Genet 2006; 70:623-30. PMID: 16907708
- Chen K, Manga P, Orlow SJ. Pink-eyed dilution protein controls the processing of tyrosinase. Mol Biol Cell 2002; 13: 1953-64. PMID: 12058062
- Gronskov K, Ek J, Sand A, Scheller R, Bygum A, Brixen K, Brondum-Nielsen K, Rosenberg T. Birth prevalence and mutation spectrum in Danish patients with autosomal recessive albinism. Invest Ophthalmol Vis Sci 2009; 50: 1058-64. PMID: 19060277
- Hutton SM, Spritz RA. A comprehensive genetic study of autosomal recessive ocular albinism in Caucasian patients. Invest Ophthalmol Vis Sci 2008a; 49: 868-72. PMID: 18326704
- Hutton SM, Spritz RA. Comprehensive Analysis of Oculocutaneous Albinism among Non-Hispanic Caucasians Shows that OCA1 Is the Most Prevalent OCA Type. J Invest Dermatol 2008b; 128: 2442-50. PMID: 18463683
- Oetting WS, Fryer JP, King RA. Mutations of the human tyrosinase gene associated with tyrosinase related oculocutaneous albinism (OCA1). Mutations in brief no. 204. Online. Hum Mutat 1998; 12: 433-4. PMID: 10671066
- Rooryck C, Morice-Picard F, El?ioglu NH, Lacombe D, Taieb A, Arveiler B. Molecular diagnosis of oculocutaneous albinism: new mutations in the OCA1-4 genes and practical aspects. Pigment Cell Melanoma Res. 2008; 21: 583-7. PMID: 18821858
- Spritz RA, Ho L, Furumura M, Hearing VJ Jr. Mutational analysis of copper binding by human tyrosinase. J Invest Dermatol 1997; 109: 207-12. PMID: 9242509
- Toyofuku K, Valencia JC, Kushimoto T, Costin GE, Virador VM, Vieira WD, Ferrans VJ, Hearing VJ. The etiology of oculocutaneous albinism (OCA) type II: the pink protein modulates the processing and transport of tyrosinase. Pigment Cell Res 2002; 15: 217-24. PMID: 12028586
- Wang Y, Guo XL, Li W, Lian S. Four novel mutations of TYR gene in Chinese OCA1 patients. J Dermatol Sci 2009; 53: 80-81.PMID: 18701257
- Watabe H, Valencia JC, Yasumoto K, Kushimoto T, Ando H, Muller J, Vieira WD, Mizoguchi M, Appella E, Hearing VJ. Regulation of tyrosinase processing and trafficking by organellar pH and by proteasome activity. J Biol Chem 2004; 279: 7971-81. PMID: 14634018
- Wei A, Wang Y, Long Y, Wang Y, Guo X, Zhou Z, Zhu W, Liu J, Bian X, Lian S, Li W. A comprehensive analysis reveals mutational spectra and common alleles in chinese patients with oculocutaneous albinism. J Invest Dermatol 2010; 130: 716-24. PMID: 19865097
- Wu H, Park HY. Protein kinase C-beta-mediated complex formation between tyrosinase and TRP-1. Biochem Biophys Res Commun 2003; 311: 948-53. PMID: 14623273
EDIT HISTORY:
Created by Wei Li & Jonathan Bourne: 07/26/2004
Updated by Wei Li: 04/06/2006
Updated by Wei Li: 08/08/2007
Updated by Wei Li: 08/20/2008
Updated by Wei Li: 02/18/2009
Updated by Wei Li: 03/15/2010