GENOMIC
Mapping
3P14.1. View the map and BAC clones (data from UCSC genome browser).
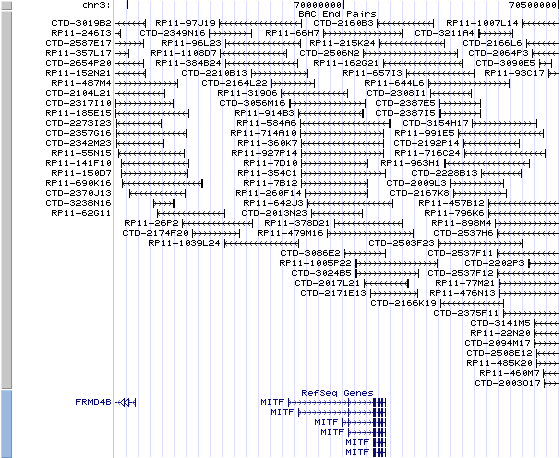
Structure
(assembly 03/2006)
isoform 1 (NM_198159): 10 exons, 228,854 bp, chr3:69871323-70100176.
isoform 2 (NM_198177): 10 exons, 102,045 bp, chr3:69998132-70100176.
isoform 3 (NM_006722): 10 exons, 204,525 bp, chr3:69895652-70100176.
isoform 4 (NM_000248): 9 exons, 31,734 bp, chr3:70068443-70100176.
isoform 5 (NM_198158): 9 exons, 31,734 bp, chr3:70068443-70100176.
isoform 6 (NM_198178): 9 exons, 89,231 bp, chr3:70010946-70100176.
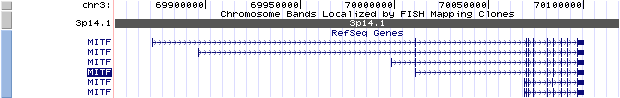
The exon 1 of six isoforms are aligned as 1A-1J-1C-1MC-1E-1H-1D-1B-1M, except 1J, 1E and 1D, each exon is associated with a distinct mRNA isoform (also denoted as isoform A(1), C(2), MC(3), H(4), B(5), M(6)), and the common exons are 2-9. All of the isoforms also produce alternative splice forms modifying exon 6 (6A/6B) that lead to inclusion or exclusion of the sequence ACIFPT upstream of the basic domain. The bHLH-Zip domain is contributed by part of exon 6B, all of exon 7 and 8, and part of exon 9.
The figure below shows the structure of the MITF gene (data from UCSC genome browser).
Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Mitf, seq2=human MITF) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
a) isoform 1( NM_198159), 4,788 bp, view ORF and the alignment to genomic.
b) isoform 2 ( NM_198177), 4,624 bp, view ORF and the alignment to genomic.
c) isoform 3 ( NM_006722), 4,700 bp, view ORF and the alignment to genomic.
d) isoform 4 (NM_000248), 4,490 bp, view ORF and the alignment to genomic.
e) isoform 5 (NM_198158), 4,472 bp, view ORF and the alignment to genomic.
f) isoform 6 (NM_198178), 4,597 bp, view ORF and the alignment to genomic.
Expression Pattern
MITF is expressed in developing NC-derived melanocyte precursors before the initial expression of Dopachrome tautomerase (DCT). M-MITF is a major isoform in NC stem cell-derived melanocytes. Isoform M is exclusively expressed in melanocytes and melanoma cells. Isoform A and isoform H are widely expressed in many cell types including melanocytes and retinal pigment epithelium (RPE). Isoform C is expressed in many cell types including RPE but not in melanocyte-lineage cells. Mitf regulation in the developing eye is isoform-selective, both temporally and spatially (Bharti, et al). Expression of mature miR-137 in melanoma cell lines down-regulates MITF expression (Bemis, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
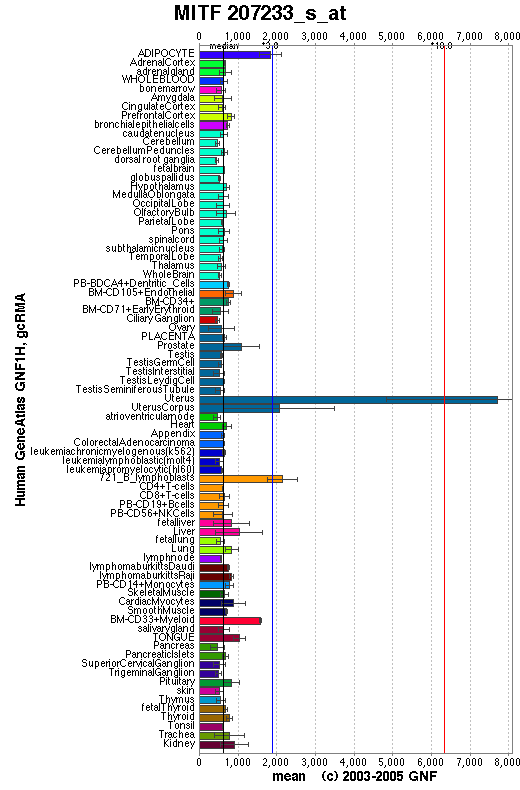
View more information about MITF in Entrez Gene, or UCSC Gene Sorter, or GeneCards.
PROTEIN
Sequence
a) microphthalmia-associated transcription factor isoform 1 (NP_937802): 520 aa, UniProtKB/Swiss-Prot entry O75030.
b) microphthalmia-associated transcription factor isoform 2 (NP_937820): 504 aa, UniProtKB/Swiss-Prot entry O75030-8.
c) microphthalmia-associated transcription factor isoform 3 (NP_006713): 519 aa, UniProtKB/Swiss-Prot entry Q8WYR3.
d) microphthalmia-associated transcription factor isoform 4 (NP_000239): 419 aa, UniProtKB/Swiss-Prot entry Q96EB5.
e) microphthalmia-associated transcription factor isoform 5 (NP_937801): 413 aa, UniProtKB/Swiss-Prot entry Q96EB5.
f) microphthalmia-associated transcription factor isoform 6 (NP_937821): 495 aa, UniProtKB/Swiss-Prot entry O75030-4.
Ortholog
Species
Chimpanzee Dog Mouse Rat Chicken GeneView
MITF
MITF
Mitf
Mitf
CMI9
Protein
XP_001138876 (520 aa)
XP_855594 (419 aa)
NP_032627 (419 aa)
XP_001065702 (520 aa)
NP_990360 (468 aa)
Identities
518/520 (99%) 392/409 (95%) 378/409 (92%) 489/520 (94%) 394/431 (91%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain (Isoform 1)
(1) Domains predicted by SMART:
a) low complexity: 41-55
b) coiled coil: 60-90
c) HLH: 311-364
d) low complexity: 435-450
e) low complexity: 504-517
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 99 to 102 NVSV.
Site : 424 to 427 NCSQ.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 507 to 510 RRSS.
c) Protein kinase C phosphorylation site:
Site : 54 to 56 TSR.
Site : 505 to 507 SSR.
Site : 506 to 508 SRR.
d) Casein kinase II phosphorylation site:
Site : 23 to 26 TYYE.
Site : 36 to 39 SSAE.
Site : 121 to 124 THLE.
Site : 189 to 192 SNCE.
Site : 236 to 239 SYNE.
Site : 293 to 296 TESE.
Site : 440 to 443 TTLD.
Site : 457 to 460 TGTE.
Site : 473 to 476 SKLE.
Site : 512 to 515 SMEE.
e) N-myristoylation site:
Site : 274 to 279 GLTISN.
Site : 458 to 463 GTEANQ.
Site : 500 to 505 GASKTS.
f) Leucine zipper pattern:
Site : 368 to 389 LENRQKKLEHANRHLLLRIQEL.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1)
ModBase predicted 3D structure from UCSC Genome Sorter:
O75030 ( isoform 1 ):
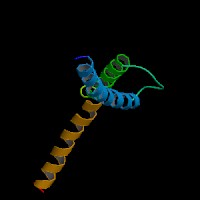
 From left to right: Front, Top, and Side views of predicted protein.
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=58,162.7Da, pI=5.93 (Isoform 1).
FUNCTION
Ontology
(1) Biological process: DNA binding, DNA-dependent regulation of transcription, sensory perception of sound.
(2) Transcription factor activity.
(3) Sequence-specific (E-boxes) (5'-CACGTG-3') DNA binding.
(4) Plays a critical role in the differentiation of various cell types as neural crest-derived melanocytes, mast cells, osteoclasts and optic cup-derived retinal pigment epithelium.
Location
Nucleus.
Interaction
Transcription factor for tyrosinase and tyrosinase-related protein 1. Efficient DNA binding requires dimerization with another bHLH protein. Binds DNA in the form of homodimer or heterodimer with either TFE3, TFEB or TFEC. Phosphorylation at Ser-405 significantly enhances the ability to bind the tyrosinase promoter.
Interactions in HPRD.
View co-occured partners in literature searched by PPI Finder.
Pathway
MITF is involved in Wnt receptor signaling pathway. MITF regulates tyrosinase and c-KIT gene. Melanogenesis pathway and Melanoma pathway in KEGG. BioCarta from NCI Cancer Genome Anatomy Project
- Melanocyte Development and Pigmentation Pathway.
MUTATION
Allele or SNP
11 mutations deposited in HGMD.
SNPs deposited in dbSNP.
Selected allelic examples described in OMIM.
Distribution
To date, 14 MITF mutations have been reported: 6 missense/nonsense, 3 splicing, 4 small deletions, and 1 gross deletion. Except the mutations deposited in HGMD, novel mutations are listed here.
Schwarzbraun, et al (2007) reported the entire MITF gene deletion in a WS2A patient with 3p deletion.
Chen, et al (2008) reported a novel frameshift mutation in a Chinese WS2 family, c.639delA.
Monma, et al (2008) identified a novel MITF mutation of IVS4 -1G>C in a Japanese family with WS2 in isoform M.
Note that the numbering of the mutations in HGMD is based on MITF isoform 4 cDNA (NM_000248), view ORF here.
Effect
Amplification of MITF gene could be detected in melanoma and other malignancies.
PHENOTYPE
Defects in MITF are the cause of Waardenburg syndrome type 2A (WS2A) [OMIM:193510]. It is a dominant inherited disorder characterized by sensorineural hearing loss and patches of depigmentation. The features show variable expression and penetrance.
Defects in MITF are a cause of Waardenburg syndrome type 2 with ocular albinism (WS2-OA)
[OMIM:103470]. It is an ocular albinism with sensorineural deafness.
Defects in MITF are the cause of Tietz syndrome [MIM:103500]. It is an autosomal dominant disorder characterized by generalized hypopigmentation and profound, congenital, bilateral deafness. Penetrance is complete.
MITF is amplified in 15% to 20% of metastatic melanomas. MITF gene amplification was shown to be associated with a reduced survival in metastatic melanoma patients, and reduction of MITF activity was shown to sensitize melanoma cell lines to chemotherapeutics (Ugurel, et al).
(Ho, et al).
REFERENCE
- Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, Erickson PF, Shellman YG, Robinson WA. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res 2008; 68:1362-8. PMID: 18316599
- Bharti K, Liu W, Csermely T, Bertuzzi S, Arnheiter H. Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development 2008; 135:1169-78. PMID: 18272592
- Chen J, Yang SZ, Liu J, Han B, Wang GJ, Zhang X, Kang DY, Dai P, Young WY, Yuan HJ. Mutation screening of MITF gene in patients with Waardenburg syndrome type 2. Yi Chuan 2008; 30:433-8. Chinese. PMID: 18424413
- Monma F, Hozumi Y, Kawaguchi M, Katagiri Y, Watanabe T, Yoshihiko I, Suzuki T. A novel MITF splice site mutation in a family with Waardenburg syndrome. J Dermatol Sci 2008; 52: 64-6. No abstract available. PMID: 18595666
- Schwarzbraun T, Ofner L, Gillessen-Kaesbach G, Schaperdoth B, Preisegger KH, Windpassinger C, Wagner K, Petek E, Kroisel PM. A new 3p interstitial deletion including the entire MITF gene causes a variation of Tietz/Waardenburg type IIA syndromes. Am J Med Genet A 2007; 143:619-24. No abstract available. PMID: 17318840
- Ugurel S, Houben R, Schrama D, Voigt H, Zapatka M, Schadendorf D, Br?cker EB, Becker JC. Microphthalmia-associated transcription factor gene amplification in metastatic melanoma is a prognostic marker for patient survival, but not a predictive marker for chemosensitivity and chemotherapy response. Clin Cancer Res 2007; 13:6344-50. PMID: 17975146
| Species | Chimpanzee | Dog | Mouse | Rat | Chicken |
| GeneView | MITF | MITF | Mitf | Mitf | CMI9 |
| Protein | XP_001138876 (520 aa) | XP_855594 (419 aa) | NP_032627 (419 aa) | XP_001065702 (520 aa) | NP_990360 (468 aa) |
| Identities | 518/520 (99%) | 392/409 (95%) | 378/409 (92%) | 489/520 (94%) | 394/431 (91%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain (Isoform 1)
(1) Domains predicted by SMART:
a) low complexity: 41-55
b) coiled coil: 60-90
c) HLH: 311-364
d) low complexity: 435-450
e) low complexity: 504-517
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 99 to 102 NVSV.
Site : 424 to 427 NCSQ.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 507 to 510 RRSS.
c) Protein kinase C phosphorylation site:
Site : 54 to 56 TSR.
Site : 505 to 507 SSR.
Site : 506 to 508 SRR.
d) Casein kinase II phosphorylation site:
Site : 23 to 26 TYYE.
Site : 36 to 39 SSAE.
Site : 121 to 124 THLE.
Site : 189 to 192 SNCE.
Site : 236 to 239 SYNE.
Site : 293 to 296 TESE.
Site : 440 to 443 TTLD.
Site : 457 to 460 TGTE.
Site : 473 to 476 SKLE.
Site : 512 to 515 SMEE.
e) N-myristoylation site:
Site : 274 to 279 GLTISN.
Site : 458 to 463 GTEANQ.
Site : 500 to 505 GASKTS.
f) Leucine zipper pattern:
Site : 368 to 389 LENRQKKLEHANRHLLLRIQEL.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1)
ModBase predicted 3D structure from UCSC Genome Sorter:
O75030 ( isoform 1 ):


 From left to right: Front, Top, and Side views of predicted protein.
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=58,162.7Da, pI=5.93 (Isoform 1).
(1) Domains predicted by SMART:
a) low complexity: 41-55
b) coiled coil: 60-90
c) HLH: 311-364
d) low complexity: 435-450
e) low complexity: 504-517
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 99 to 102 NVSV.
Site : 424 to 427 NCSQ.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 507 to 510 RRSS.
c) Protein kinase C phosphorylation site:
Site : 54 to 56 TSR.
Site : 505 to 507 SSR.
Site : 506 to 508 SRR.
d) Casein kinase II phosphorylation site:
Site : 23 to 26 TYYE.
Site : 36 to 39 SSAE.
Site : 121 to 124 THLE.
Site : 189 to 192 SNCE.
Site : 236 to 239 SYNE.
Site : 293 to 296 TESE.
Site : 440 to 443 TTLD.
Site : 457 to 460 TGTE.
Site : 473 to 476 SKLE.
Site : 512 to 515 SMEE.
e) N-myristoylation site:
Site : 274 to 279 GLTISN.
Site : 458 to 463 GTEANQ.
Site : 500 to 505 GASKTS.
f) Leucine zipper pattern:
Site : 368 to 389 LENRQKKLEHANRHLLLRIQEL.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure from UCSC Genome Sorter:
O75030 ( isoform 1 ):


 From left to right: Front, Top, and Side views of predicted protein.
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=58,162.7Da, pI=5.93 (Isoform 1).
FUNCTION
Ontology
(1) Biological process: DNA binding, DNA-dependent regulation of transcription, sensory perception of sound.
(2) Transcription factor activity.
(3) Sequence-specific (E-boxes) (5'-CACGTG-3') DNA binding.
(4) Plays a critical role in the differentiation of various cell types as neural crest-derived melanocytes, mast cells, osteoclasts and optic cup-derived retinal pigment epithelium.
Location
Nucleus.
Interaction
Transcription factor for tyrosinase and tyrosinase-related protein 1. Efficient DNA binding requires dimerization with another bHLH protein. Binds DNA in the form of homodimer or heterodimer with either TFE3, TFEB or TFEC. Phosphorylation at Ser-405 significantly enhances the ability to bind the tyrosinase promoter.
Interactions in HPRD.
View co-occured partners in literature searched by PPI Finder.
Pathway
MITF is involved in Wnt receptor signaling pathway. MITF regulates tyrosinase and c-KIT gene. Melanogenesis pathway and Melanoma pathway in KEGG. BioCarta from NCI Cancer Genome Anatomy Project - Melanocyte Development and Pigmentation Pathway.
MUTATION
Allele or SNP
11 mutations deposited in HGMD.
SNPs deposited in dbSNP.
Selected allelic examples described in OMIM.
Distribution
To date, 14 MITF mutations have been reported: 6 missense/nonsense, 3 splicing, 4 small deletions, and 1 gross deletion. Except the mutations deposited in HGMD, novel mutations are listed here.
Schwarzbraun, et al (2007) reported the entire MITF gene deletion in a WS2A patient with 3p deletion.
Chen, et al (2008) reported a novel frameshift mutation in a Chinese WS2 family, c.639delA.
Monma, et al (2008) identified a novel MITF mutation of IVS4 -1G>C in a Japanese family with WS2 in isoform M.
Note that the numbering of the mutations in HGMD is based on MITF isoform 4 cDNA (NM_000248), view ORF here.
Effect
Amplification of MITF gene could be detected in melanoma and other malignancies.
PHENOTYPE
Defects in MITF are the cause of Waardenburg syndrome type 2A (WS2A) [OMIM:193510]. It is a dominant inherited disorder characterized by sensorineural hearing loss and patches of depigmentation. The features show variable expression and penetrance.
Defects in MITF are a cause of Waardenburg syndrome type 2 with ocular albinism (WS2-OA) [OMIM:103470]. It is an ocular albinism with sensorineural deafness.
Defects in MITF are the cause of Tietz syndrome [MIM:103500]. It is an autosomal dominant disorder characterized by generalized hypopigmentation and profound, congenital, bilateral deafness. Penetrance is complete.
MITF is amplified in 15% to 20% of metastatic melanomas. MITF gene amplification was shown to be associated with a reduced survival in metastatic melanoma patients, and reduction of MITF activity was shown to sensitize melanoma cell lines to chemotherapeutics (Ugurel, et al).
(Ho, et al).
REFERENCE
- Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, Erickson PF, Shellman YG, Robinson WA. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res 2008; 68:1362-8. PMID: 18316599
- Bharti K, Liu W, Csermely T, Bertuzzi S, Arnheiter H. Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development 2008; 135:1169-78. PMID: 18272592
- Chen J, Yang SZ, Liu J, Han B, Wang GJ, Zhang X, Kang DY, Dai P, Young WY, Yuan HJ. Mutation screening of MITF gene in patients with Waardenburg syndrome type 2. Yi Chuan 2008; 30:433-8. Chinese. PMID: 18424413
- Monma F, Hozumi Y, Kawaguchi M, Katagiri Y, Watanabe T, Yoshihiko I, Suzuki T. A novel MITF splice site mutation in a family with Waardenburg syndrome. J Dermatol Sci 2008; 52: 64-6. No abstract available. PMID: 18595666
- Schwarzbraun T, Ofner L, Gillessen-Kaesbach G, Schaperdoth B, Preisegger KH, Windpassinger C, Wagner K, Petek E, Kroisel PM. A new 3p interstitial deletion including the entire MITF gene causes a variation of Tietz/Waardenburg type IIA syndromes. Am J Med Genet A 2007; 143:619-24. No abstract available. PMID: 17318840
- Ugurel S, Houben R, Schrama D, Voigt H, Zapatka M, Schadendorf D, Br?cker EB, Becker JC. Microphthalmia-associated transcription factor gene amplification in metastatic melanoma is a prognostic marker for patient survival, but not a predictive marker for chemosensitivity and chemotherapy response. Clin Cancer Res 2007; 13:6344-50. PMID: 17975146