GENOMIC
Mapping
13q22.3. View the map and BAC clones (data from UCSC genome browser).
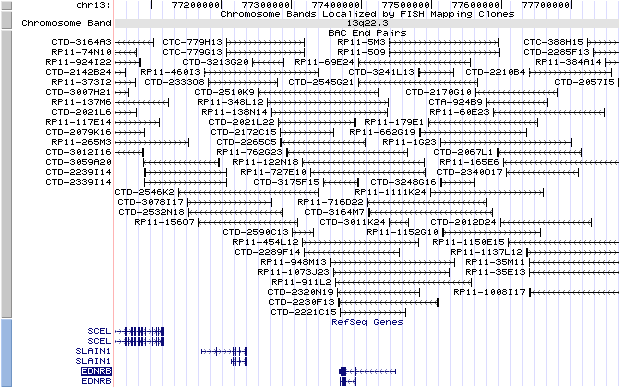
Structure
(assembly 03/2006)
EDNRB isoform 1 (NM_000115): 8 exons, 80,049 bp, chr13:77367617-77447665.
EDNRB isoform 2 (NM_003991): 7 exons, 22,392 bp, chr13:77368544-77390935.
Variant (2) differs in the 5' UTR, 3' UTR, and 3' coding sequence compared to variant (1). The resulting isoform 2 has a shorter and distinct C-terminus compared to isoform 1.
The figure below shows the structure of the EDNRB gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Ednrb, seq2=human EDNRB) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
a) EDNRB isoform 1 (NM_000115), 4,286 bp, view ORF and the alignment to genomic.
b) EDNRB isoform 2 (NM_003991), 1,578 bp, view ORF and the alignment to genomic.
Expression Pattern
Expressed in placental stem villi vessels, but not in cultured placental villi smooth muscle cells.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
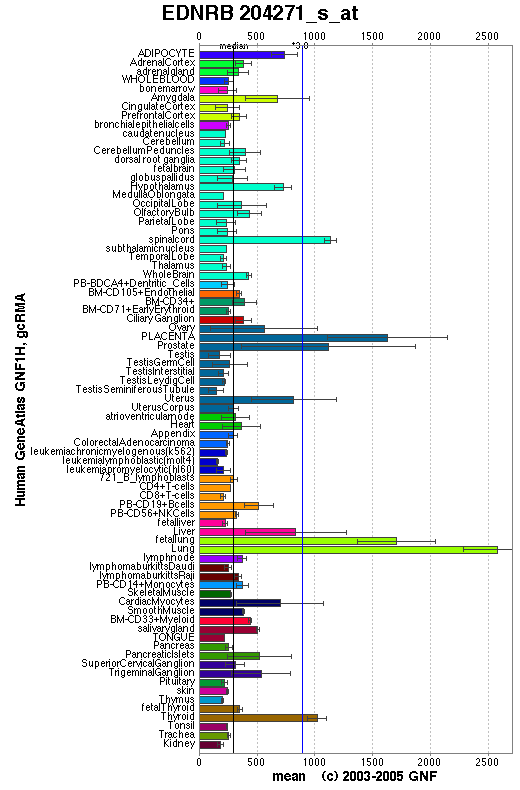
View more information in Entrez Gene, or UCSC Gene Sorter, or GeneCards.
PROTEIN
Sequence
a) endothelin receptor type B isoform 1 (NP_000106): 442 aa, UniProtKB/Swiss-Prot entry P24530.
b) endothelin receptor type B isoform 2 (NP_003982): 436 aa, UniProtKB/Swiss-Prot entry Q8NHM6.
Ortholog
Species
Chimpanzee Dog Mouse Rat Fowl GeneView
EDNRB
EDNRB
Ednrb
Ednrb
EDNRB
Protein
XP_509693 (532 aa)
NP_001010943 (442 aa)
NP_031930 (442 aa)
NP_059029 (442 aa)
XP_41700 (560 aa)
Identities
442/442 (100%) 430/442 (97%) 393/443 (88%) 393/443 (88%) 345/414 (83%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain (isoform 1)
(1) Domains predicted by SMART:
a) signal peptide: 1-26
b) Pfam:7tm_1: 118-386
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a MEMBRANE PROTEIN
which have 8 transmembrane helices.
No. N terminal transmembrane region C terminal type length 1 5 PSLCGRALVALVLACGLSRIWGE 27 SECONDARY 23 2 100 FKYINTVVSCLVFVLGIIGNSTL 122 PRIMARY 23 3 138 ILIASLALGDLLHIVIDIPINVY 160 SECONDARY 23 4 173 MCKLVPFIQKASVGITVLSLCAL 195 SECONDARY 23 5 221 EIVLIWVVSVVLAVPEAIGFDII 243 PRIMARY 23 6 277 LFSFYFCLPLAITAFFYTLMTCE 299 SECONDARY 23 7 325 VFCLVLVFALCWLPLHLSRILKL 347 PRIMARY 23 8 358 CELLSFLLVLDYIGINMASLNSC 380 SECONDARY 23
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 59 to 62 NASL.
Site : 119 to 122 NSTL.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 302 to 305 RKKS.
c) Protein kinase C phosphorylation site:
Site : 99 to 101 TFK.
Site : 271 to 273 TAK.
Site : 390 to 392 SKR.
Site : 436 to 438 SNK.
d) Casein kinase II phosphorylation site:
Site : 218 to 221 TAVE.
Site : 271 to 274 TAKD.
Site : 407 to 410 SFEE.
Site : 413 to 416 SLEE.
e)Tyrosine kinase phosphorylation site:
Site : 424 to 430 KANDHGY.
f) N-myristoylation site:
Site : 57 to 62 GSNASL.
Site : 115 to 120 GIIGNS.
Site : 170 to 175 GAEMCK.
Site : 306 to 311 GMQIAL.
Site : 371 to 376 GINMAS.
g) G-protein coupled receptors family 1 signature:
Site : 187 to 203 ITVLSLCALSIDRYRAV.
Note that palmitoylation of Cys-402 was confirmed by the palmitoylation of Cys-402 in a deletion mutant lacking both Cys-403 and Cys-405.
(2) Predicted results of subprograms by PSORT II:
a) Seems to have a cleavable signal peptide (1 to 26)
b) Tentative number of TMS(s) for the threshold 0.5: 7
INTEGRAL Likelihood = -4.88 Transmembrane 102 - 118
INTEGRAL Likelihood = -3.45 Transmembrane 137 - 153
INTEGRAL Likelihood = -0.37 Transmembrane 179 - 195
INTEGRAL Likelihood = -8.92 Transmembrane 218 - 234
INTEGRAL Likelihood = -2.02 Transmembrane 276 - 292
INTEGRAL Likelihood = -8.07 Transmembrane 325 - 341
INTEGRAL Likelihood = -0.80 Transmembrane 360 - 376
PERIPHERAL Likelihood = 8.01 (at 377)
ALOM score: -8.92 (number of TMSs: 7)
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: KYSS
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
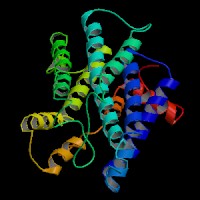
3D Model
(1)
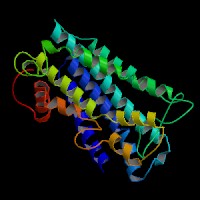
ModBase predicted 3D structure from UCSC Gene Sorter:
P24530 (endothelin receptor type B isoform 1):
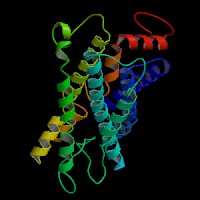
Q8NHM6 (endothelin receptor type B isoform 2):
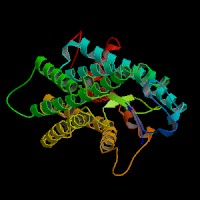
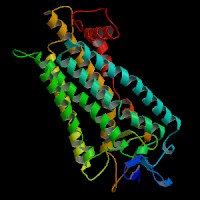
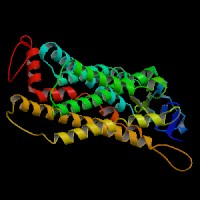
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=49,644Da, pI=9.15 (isoform 1).
FUNCTION
Ontology
(1) Biological process: neural crest cell migration, signal transduction, smooth muscle contraction, sensory perception of sound.
(2) Regulation of blood pressure.
(3) G-protein coupled receptor activity, rhodopsin-like.
(4) Inositol phosphate-mediated signaling.
(5) Melanocyte differentiation.
Location
Plasma membrane.
Interaction
The protein encoded by this gene is a G protein-coupled receptor which activates a phosphatidylinositol-calcium second messenger system. Its ligand, endothelin, consists of a family of three potent vasoactive peptides: ET1, ET2, and ET3. While both isoforms bind ET1, they exhibit different responses upon binding, suggesting that they may be functionally distinct. The interaction of this endothelin with EDNRB is essential for development of neural crest-derived cell lineages, such as melanocytes and enteric neurons.
View interactions in HPRD
View co-occured partners in literature searched by PPI Finder.
Pathway
Neuroactive ligand-receptor interaction, Melanogenesis in KEGG.
MUTATION
Allele or SNP
26 mutations deposited in HGMD.
SNPs deposited in dbSNP.
Selected allelic examples described in OMIM.
Distribution
Except the mutations deposited in HGMD, other novel mutations of EDNRB are liste here.
(1) One novel missense mutation, V111Q, was detected in Thai Hirschsprung disease (HSCR) (Sangkhathat, et al (2006)).
(2) One novel missense mutation, V185M, was found in Chinese Hirschsprung disease (HSCR) (Zhou, et al (2006)) and another heterozygous novel missense mutation (C90R) was found in a Taiwanese HSCR patient (Chen, et al (2006)) .
Note that the numbering of the mutations in HGMD is based on EDNRB cDNA (NM_000115), view ORF here.
Effect
PHENOTYPE
Mutations in this gene and EDNRB have been associated with Hirschsprung disease (HSCR) and Waardenburg syndrome (WS), which are congenital disorders involving neural crest-derived cells.
Defects in EDNRB are a cause of Waardenburg syndrome type IV (WS4) [MIM:277580]; also known as Waardenburg-Shah syndrome. WS4 is characterized by the association of Waardenburg features (depigmentation and deafness) and the absence of enteric ganglia in the distal part of the intestine (Hirschsprung disease).
Defects in EDNRB are the cause of Hirschsprung disease type 2 (HSCR2) [MIM:600155]; also known as aganglionic megacolon (MGC). It is a congenital disorder characterized by absence of enteric ganglia along a variable length of the intestine. It is the most common cause of congenital intestinal obstruction. Early symptoms range from complete acute neonatal obstruction, characterized by vomiting, abdominal distention and failure to pass stool, to chronic constipation in the older child.
Defects in EDNRB are the cause of ABCD syndrome [MIM:600501]. ABCD syndrome is an autosomal recessive syndrome characterized by albinism, black lock at temporal occipital region, bilateral deafness, aganglionosis of the large intestine and total absence of neurocytes and nerve fibers in the small intestine.
EDNRB has been reported in association with atherosclerosis, Hirschsprung disease, migraine.
REFERENCE
- Chen WC, Chang SS, Sy ED, Tsai MC. A De Novo novel mutation of the EDNRB gene in a Taiwanese boy with Hirschsprung disease. J Formos Med Assoc 2006; 105:349-54. PMID: 16618617
- Sangkhathat S, Kusafuka T, Chengkriwate P, Patrapinyokul S, Sangthong B, Fukuzawa M. Mutations and polymorphisms of Hirschsprung disease candidate genes in Thai patients. J Hum Genet 2006; 51:1126-32. PMID: 17009072
- Zhou MN, Li JC, Ding SP. [Coding-sequence mutation and polymorphism analysis of EDNRB gene in patients with Hirschsprung's disease from Zhejiang region]. Fen Zi Xi Bao Sheng Wu Xue Bao 2006; 39:61-5. Chinese. PMID: 16944573
| Species | Chimpanzee | Dog | Mouse | Rat | Fowl |
| GeneView | EDNRB | EDNRB | Ednrb | Ednrb | EDNRB |
| Protein | XP_509693 (532 aa) | NP_001010943 (442 aa) | NP_031930 (442 aa) | NP_059029 (442 aa) | XP_41700 (560 aa) |
| Identities | 442/442 (100%) | 430/442 (97%) | 393/443 (88%) | 393/443 (88%) | 345/414 (83%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain (isoform 1)
(1) Domains predicted by SMART:
a) signal peptide: 1-26
b) Pfam:7tm_1: 118-386
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a MEMBRANE PROTEIN
which have 8 transmembrane helices.
No. N terminal transmembrane region C terminal type length 1 5 PSLCGRALVALVLACGLSRIWGE 27 SECONDARY 23 2 100 FKYINTVVSCLVFVLGIIGNSTL 122 PRIMARY 23 3 138 ILIASLALGDLLHIVIDIPINVY 160 SECONDARY 23 4 173 MCKLVPFIQKASVGITVLSLCAL 195 SECONDARY 23 5 221 EIVLIWVVSVVLAVPEAIGFDII 243 PRIMARY 23 6 277 LFSFYFCLPLAITAFFYTLMTCE 299 SECONDARY 23 7 325 VFCLVLVFALCWLPLHLSRILKL 347 PRIMARY 23 8 358 CELLSFLLVLDYIGINMASLNSC 380 SECONDARY 23
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 59 to 62 NASL.
Site : 119 to 122 NSTL.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 302 to 305 RKKS.
c) Protein kinase C phosphorylation site:
Site : 99 to 101 TFK.
Site : 271 to 273 TAK.
Site : 390 to 392 SKR.
Site : 436 to 438 SNK.
d) Casein kinase II phosphorylation site:
Site : 218 to 221 TAVE.
Site : 271 to 274 TAKD.
Site : 407 to 410 SFEE.
Site : 413 to 416 SLEE.
e)Tyrosine kinase phosphorylation site:
Site : 424 to 430 KANDHGY.
f) N-myristoylation site:
Site : 57 to 62 GSNASL.
Site : 115 to 120 GIIGNS.
Site : 170 to 175 GAEMCK.
Site : 306 to 311 GMQIAL.
Site : 371 to 376 GINMAS.
g) G-protein coupled receptors family 1 signature:
Site : 187 to 203 ITVLSLCALSIDRYRAV.
Note that palmitoylation of Cys-402 was confirmed by the palmitoylation of Cys-402 in a deletion mutant lacking both Cys-403 and Cys-405.
(2) Predicted results of subprograms by PSORT II:
a) Seems to have a cleavable signal peptide (1 to 26)
b) Tentative number of TMS(s) for the threshold 0.5: 7
INTEGRAL Likelihood = -4.88 Transmembrane 102 - 118
INTEGRAL Likelihood = -3.45 Transmembrane 137 - 153
INTEGRAL Likelihood = -0.37 Transmembrane 179 - 195
INTEGRAL Likelihood = -8.92 Transmembrane 218 - 234
INTEGRAL Likelihood = -2.02 Transmembrane 276 - 292
INTEGRAL Likelihood = -8.07 Transmembrane 325 - 341
INTEGRAL Likelihood = -0.80 Transmembrane 360 - 376
PERIPHERAL Likelihood = 8.01 (at 377)
ALOM score: -8.92 (number of TMSs: 7)
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: KYSS
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1)
ModBase predicted 3D structure from UCSC Gene Sorter:
P24530 (endothelin receptor type B isoform 1):



Q8NHM6 (endothelin receptor type B isoform 2):



From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=49,644Da, pI=9.15 (isoform 1).
(1) Domains predicted by SMART:
a) signal peptide: 1-26
b) Pfam:7tm_1: 118-386
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a MEMBRANE PROTEIN
which have 8 transmembrane helices.
| No. | N terminal | transmembrane region | C terminal | type | length |
| 1 | 5 | PSLCGRALVALVLACGLSRIWGE | 27 | SECONDARY | 23 |
| 2 | 100 | FKYINTVVSCLVFVLGIIGNSTL | 122 | PRIMARY | 23 |
| 3 | 138 | ILIASLALGDLLHIVIDIPINVY | 160 | SECONDARY | 23 |
| 4 | 173 | MCKLVPFIQKASVGITVLSLCAL | 195 | SECONDARY | 23 |
| 5 | 221 | EIVLIWVVSVVLAVPEAIGFDII | 243 | PRIMARY | 23 |
| 6 | 277 | LFSFYFCLPLAITAFFYTLMTCE | 299 | SECONDARY | 23 |
| 7 | 325 | VFCLVLVFALCWLPLHLSRILKL | 347 | PRIMARY | 23 |
| 8 | 358 | CELLSFLLVLDYIGINMASLNSC | 380 | SECONDARY | 23 |
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 59 to 62 NASL.
Site : 119 to 122 NSTL.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 302 to 305 RKKS.
c) Protein kinase C phosphorylation site:
Site : 99 to 101 TFK.
Site : 271 to 273 TAK.
Site : 390 to 392 SKR.
Site : 436 to 438 SNK.
d) Casein kinase II phosphorylation site:
Site : 218 to 221 TAVE.
Site : 271 to 274 TAKD.
Site : 407 to 410 SFEE.
Site : 413 to 416 SLEE.
e)Tyrosine kinase phosphorylation site:
Site : 424 to 430 KANDHGY.
f) N-myristoylation site:
Site : 57 to 62 GSNASL.
Site : 115 to 120 GIIGNS.
Site : 170 to 175 GAEMCK.
Site : 306 to 311 GMQIAL.
Site : 371 to 376 GINMAS.
g) G-protein coupled receptors family 1 signature:
Site : 187 to 203 ITVLSLCALSIDRYRAV.
Note that palmitoylation of Cys-402 was confirmed by the palmitoylation of Cys-402 in a deletion mutant lacking both Cys-403 and Cys-405.
(2) Predicted results of subprograms by PSORT II:
a) Seems to have a cleavable signal peptide (1 to 26)
b) Tentative number of TMS(s) for the threshold 0.5: 7
INTEGRAL Likelihood = -4.88 Transmembrane 102 - 118
INTEGRAL Likelihood = -3.45 Transmembrane 137 - 153
INTEGRAL Likelihood = -0.37 Transmembrane 179 - 195
INTEGRAL Likelihood = -8.92 Transmembrane 218 - 234
INTEGRAL Likelihood = -2.02 Transmembrane 276 - 292
INTEGRAL Likelihood = -8.07 Transmembrane 325 - 341
INTEGRAL Likelihood = -0.80 Transmembrane 360 - 376
PERIPHERAL Likelihood = 8.01 (at 377)
ALOM score: -8.92 (number of TMSs: 7)
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: KYSS
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure from UCSC Gene Sorter:
P24530 (endothelin receptor type B isoform 1):



Q8NHM6 (endothelin receptor type B isoform 2):



2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=49,644Da, pI=9.15 (isoform 1).
FUNCTION
Ontology
(1) Biological process: neural crest cell migration, signal transduction, smooth muscle contraction, sensory perception of sound.
(2) Regulation of blood pressure.
(3) G-protein coupled receptor activity, rhodopsin-like.
(4) Inositol phosphate-mediated signaling.
(5) Melanocyte differentiation.
Location
Plasma membrane.
Interaction
The protein encoded by this gene is a G protein-coupled receptor which activates a phosphatidylinositol-calcium second messenger system. Its ligand, endothelin, consists of a family of three potent vasoactive peptides: ET1, ET2, and ET3. While both isoforms bind ET1, they exhibit different responses upon binding, suggesting that they may be functionally distinct. The interaction of this endothelin with EDNRB is essential for development of neural crest-derived cell lineages, such as melanocytes and enteric neurons.
View interactions in HPRD
View co-occured partners in literature searched by PPI Finder.
Pathway
Neuroactive ligand-receptor interaction, Melanogenesis in KEGG.
MUTATION
Allele or SNP
26 mutations deposited in HGMD.
SNPs deposited in dbSNP.
Selected allelic examples described in OMIM.
Distribution
Except the mutations deposited in HGMD, other novel mutations of EDNRB are liste here.
(1) One novel missense mutation, V111Q, was detected in Thai Hirschsprung disease (HSCR) (Sangkhathat, et al (2006)).
(2) One novel missense mutation, V185M, was found in Chinese Hirschsprung disease (HSCR) (Zhou, et al (2006)) and another heterozygous novel missense mutation (C90R) was found in a Taiwanese HSCR patient (Chen, et al (2006)) .
Note that the numbering of the mutations in HGMD is based on EDNRB cDNA (NM_000115), view ORF here.
Effect
PHENOTYPE
Mutations in this gene and EDNRB have been associated with Hirschsprung disease (HSCR) and Waardenburg syndrome (WS), which are congenital disorders involving neural crest-derived cells.
Defects in EDNRB are a cause of Waardenburg syndrome type IV (WS4) [MIM:277580]; also known as Waardenburg-Shah syndrome. WS4 is characterized by the association of Waardenburg features (depigmentation and deafness) and the absence of enteric ganglia in the distal part of the intestine (Hirschsprung disease).
Defects in EDNRB are the cause of Hirschsprung disease type 2 (HSCR2) [MIM:600155]; also known as aganglionic megacolon (MGC). It is a congenital disorder characterized by absence of enteric ganglia along a variable length of the intestine. It is the most common cause of congenital intestinal obstruction. Early symptoms range from complete acute neonatal obstruction, characterized by vomiting, abdominal distention and failure to pass stool, to chronic constipation in the older child.
Defects in EDNRB are the cause of ABCD syndrome [MIM:600501]. ABCD syndrome is an autosomal recessive syndrome characterized by albinism, black lock at temporal occipital region, bilateral deafness, aganglionosis of the large intestine and total absence of neurocytes and nerve fibers in the small intestine.
EDNRB has been reported in association with atherosclerosis, Hirschsprung disease, migraine.
REFERENCE
- Chen WC, Chang SS, Sy ED, Tsai MC. A De Novo novel mutation of the EDNRB gene in a Taiwanese boy with Hirschsprung disease. J Formos Med Assoc 2006; 105:349-54. PMID: 16618617
- Sangkhathat S, Kusafuka T, Chengkriwate P, Patrapinyokul S, Sangthong B, Fukuzawa M. Mutations and polymorphisms of Hirschsprung disease candidate genes in Thai patients. J Hum Genet 2006; 51:1126-32. PMID: 17009072
- Zhou MN, Li JC, Ding SP. [Coding-sequence mutation and polymorphism analysis of EDNRB gene in patients with Hirschsprung's disease from Zhejiang region]. Fen Zi Xi Bao Sheng Wu Xue Bao 2006; 39:61-5. Chinese. PMID: 16944573