GENOMIC
Mapping
19p13.3. View the map and BAC contig (data from UCSC genome browser).
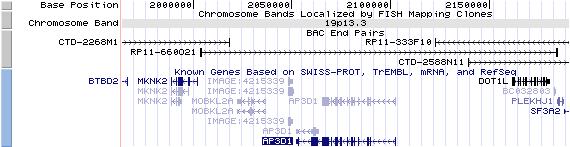
Structure
(assembly 07/03)
AP3D1/NM_003938: 30 exons, 50,567bp, Chr19: 2,051,988-2,102,554.
The figure below shows the structure of the AP3D1 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
AP3D1 (NM_003938), 4,878bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: Present in all adult tissues examined with the highest levels in skeletal muscle, heart, pancreas and testis.
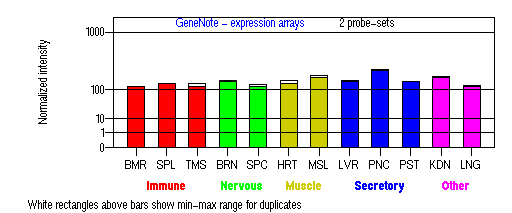
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle; LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
AP-3 complex delta subunit
(NP_003929): 1138aa, ExPaSy NiceProt view of Swiss-Prot:O14617.
Synonyms: Adaptor-related protein complex 3 delta 1 subunit, Delta-adaptin 3, Delta-adaptin.
Ortholog
| Species | Mouse | Rat | Zebrafish | Worm | Drosophila |
| GeneView | mh/Ap3d | LOC314633 | 03334 | apt-5 | g/CG10986 |
| Protein | NP_031486 (1199aa) | XP_234908 (1204aa) | 17714 (966aa) | NP_494570 (1251aa) | NP_524785 (1034aa) |
| Identities | 86%/1014aa | 86%/1015aa | 81%/748aa | 46% /570aa | 58%/539aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) low complexity: 460 - 472, 535 - 548, 659 - 677, 838 - 893.
b) coiled coil: 722 - 756.
(2) Transmembrane domains predicted by SOSUI:
1 transmembrane helix, 272-294.
(3) Pfam domains: PF01602 - Adaptin N terminal region.
(4) CDD conserved domain: KOG1059, vesicle coat complex AP-3, delta subunit (Intracellular trafficking, secretion, and vesicular transport).
(5) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) Lysine-rich region profile : [occurs frequently]
828 - 893: score=20.975
b) Tyrosine kinase phosphorylation site : [occurs frequently]
29 - 35: Ked.Eak.Y,
359 - 365: Ral.Dll.Y
c) Amidation site : [occurs frequently]
255 - 258: lGKK,
752 - 755: kGKR,
886 - 889: kGKK.
d) N-glycosylation site : [occurs frequently]
296 - 299: NHSA,
955 - 958: NRSS.
e) cAMP- and cGMP-dependent protein kinase phosphorylation site : [occurs frequently]
755 - 758: RRhS,
866 - 869: KKkS,
888 - 891: KKkS.
f) Tyrosine sulfation site : [occurs frequently]
385 - 399:
dkaegttYrdelltk,
717 - 731:
glpmsdqYvkleeer,
792 - 806:
dkdpndpYraldidl.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: found KLPI at 815
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: too long of a tail
i) Dileucine motif in the tail: found LL at 1118.
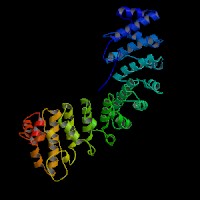
3D Model
(1) ModBase: 1 match found, results here.
(2) ModBase predicted comparative 3D structures on O14617 (data from UCSC Gene Sorter). (from left to right: Front, Top, Side view)
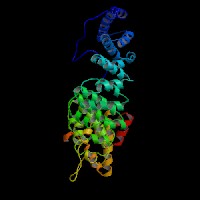
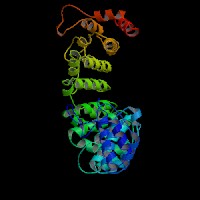
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=128,637Da, pI=8.87 (NP_003929).
(1) Domains predicted by SMART:
a) low complexity: 460 - 472, 535 - 548, 659 - 677, 838 - 893.
b) coiled coil: 722 - 756.
(2) Transmembrane domains predicted by SOSUI:
1 transmembrane helix, 272-294.
(3) Pfam domains: PF01602 - Adaptin N terminal region.
(4) CDD conserved domain: KOG1059, vesicle coat complex AP-3, delta subunit (Intracellular trafficking, secretion, and vesicular transport).
(5) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) Lysine-rich region profile : [occurs frequently]
828 - 893: score=20.975
b) Tyrosine kinase phosphorylation site : [occurs frequently]
29 - 35: Ked.Eak.Y,
359 - 365: Ral.Dll.Y
c) Amidation site : [occurs frequently]
255 - 258: lGKK,
752 - 755: kGKR,
886 - 889: kGKK.
d) N-glycosylation site : [occurs frequently]
296 - 299: NHSA,
955 - 958: NRSS.
e) cAMP- and cGMP-dependent protein kinase phosphorylation site : [occurs frequently]
755 - 758: RRhS,
866 - 869: KKkS,
888 - 891: KKkS.
f) Tyrosine sulfation site : [occurs frequently]
385 - 399:
dkaegttYrdelltk,
717 - 731:
glpmsdqYvkleeer,
792 - 806:
dkdpndpYraldidl.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: found KLPI at 815
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: too long of a tail
i) Dileucine motif in the tail: found LL at 1118.
3D Model
(1) ModBase: 1 match found, results here.
(2) ModBase predicted comparative 3D structures on O14617 (data from UCSC Gene Sorter). (from left to right: Front, Top, Side view)



2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=128,637Da, pI=8.87 (NP_003929).
FUNCTION
Ontology
a) Biological process: intracellular protein transport, vesicle-mediated transport (overview of trafficking pathway here).
b) Biological process: eye pigment biosynthesis.
c) Component of Golgi apparatus.
Location
Component of the coat surrounding the cytoplasmic face of coated vesicles located at the Golgi complex.
Interaction
AP3D1 is a subunit of the AP3 adaptor-like complex, which is not associated with clathrin. The AP-3 complex is a heterotetramer composed of two large adaptins (delta/AP3D1 and beta3A/AP3B2 or beta3B/AP3B1), a medium adaptin (mu3A/AP3M1 or mu3B/AP3M2) and a small adaptin (sigma3A/AP3S1 or sigma3B/AP3S2).
AP-3, has been identified that interacts with dileucine motifs and mediates endosomal/lysosomal transport in yeast, drosophila, and mammals. In addition, the AP3M1 subunit interacts with tyrosinase for lysosomal targeting (Honing, et al). AP-3 complex interacts with CD1 antigen presenting molecules.
Association of AP-3 with membranes in vitro was enhanced by GTPgammaS and inhibited by brefeldin A (BFA), an inhibitor of ARF1 guanine nucleotide exchange. In addition, recombinant myristoylated ARF1 promoted association of AP-3 with membranes. The membrane recruitment of AP-3 is promoted by ARF1-GTP (Ooi, et al (1998)).
6 proteins are shown to be associated with APL5 in Yeast GRID.
Ap3d drosophila homolog CG10986 interaction information in CuraGen interaction database.
Pathway
The AP3D1 subunit is implicated in intracellular biogenesis and trafficking of pigment granules and possibly platelet dense granules and neurotransmitter vesicles. AP-3 complex is associated with the Golgi region as well as more peripheral structures. It facilitates the budding of vesicles from the Golgi membrane and appears to be involved in the sorting of a subset of transmembrane proteins targeted to lysosomes and lysosome-related organelles. In yeast, AP-3 sorts the vacuolar membrane enzymes, alkaline phosphatase and Vamp3p, a vacuolar t-SNARE (Odorizzi, et al). Faundez, et al found that the AP-3 complex is involved in synaptic vesicle formation in neuronal cells. Likewise, Sugita, et al concluded that there is an AP-3 dependent pathway for antigen presentation by CD1B (human) or CD1d (mice) molecules. The mechanism of AP-3 dependent pathway is distinct from that dependent on HPS1 (Feng, et al) (view diagram of BLOC-3 and AP-3 pathway here).
a) Biological process: intracellular protein transport, vesicle-mediated transport (overview of trafficking pathway here).
b) Biological process: eye pigment biosynthesis.
c) Component of Golgi apparatus.
Location
Component of the coat surrounding the cytoplasmic face of coated vesicles located at the Golgi complex.
Interaction
AP3D1 is a subunit of the AP3 adaptor-like complex, which is not associated with clathrin. The AP-3 complex is a heterotetramer composed of two large adaptins (delta/AP3D1 and beta3A/AP3B2 or beta3B/AP3B1), a medium adaptin (mu3A/AP3M1 or mu3B/AP3M2) and a small adaptin (sigma3A/AP3S1 or sigma3B/AP3S2). AP-3, has been identified that interacts with dileucine motifs and mediates endosomal/lysosomal transport in yeast, drosophila, and mammals. In addition, the AP3M1 subunit interacts with tyrosinase for lysosomal targeting (Honing, et al). AP-3 complex interacts with CD1 antigen presenting molecules.
Association of AP-3 with membranes in vitro was enhanced by GTPgammaS and inhibited by brefeldin A (BFA), an inhibitor of ARF1 guanine nucleotide exchange. In addition, recombinant myristoylated ARF1 promoted association of AP-3 with membranes. The membrane recruitment of AP-3 is promoted by ARF1-GTP (Ooi, et al (1998)).
6 proteins are shown to be associated with APL5 in Yeast GRID.
Ap3d drosophila homolog CG10986 interaction information in CuraGen interaction database.
Pathway
The AP3D1 subunit is implicated in intracellular biogenesis and trafficking of pigment granules and possibly platelet dense granules and neurotransmitter vesicles. AP-3 complex is associated with the Golgi region as well as more peripheral structures. It facilitates the budding of vesicles from the Golgi membrane and appears to be involved in the sorting of a subset of transmembrane proteins targeted to lysosomes and lysosome-related organelles. In yeast, AP-3 sorts the vacuolar membrane enzymes, alkaline phosphatase and Vamp3p, a vacuolar t-SNARE (Odorizzi, et al). Faundez, et al found that the AP-3 complex is involved in synaptic vesicle formation in neuronal cells. Likewise, Sugita, et al concluded that there is an AP-3 dependent pathway for antigen presentation by CD1B (human) or CD1d (mice) molecules. The mechanism of AP-3 dependent pathway is distinct from that dependent on HPS1 (Feng, et al) (view diagram of BLOC-3 and AP-3 pathway here).
PHENOTYPE
(Animal Models)
Mutations in the Ap3d gene are the cause of mocha (mh) mutants (Katheti, et al (1998; 2003)), a mouse model of Hermansky-Pudlak syndrome (OMIM 607246). Ooi, et al (1997) showed that the garnet mutation in drosophila, which causes reduced pigmentation of the eyes and other tissues, results from a mutation in the fly AP3D1 homolog gene.
REFERENCE
- Faundez V, Horng JT, Kelly RB. A function for the AP3 coat complex in synaptic vesicle formation from endosomes. Cell 1998; 93: 423-32. PMID: 9590176
- Feng L, Novak EK, Hartnell LM, Bonifacino JS, Collinson LM, Swank RT. The Hermansky-Pudlak syndrome 1 (HPS1) and HPS2 genes independently contribute to the production and function of platelet dense granules, melanosomes, and lysosomes. Blood 2002; 99: 1651-8. PMID: 11861280
- Honing S, Sandoval IV, von Figura K. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J 1998; 17: 1304-14. PMID: 9482728
- Kantheti P, Diaz ME, Peden AE, Seong EE, Dolan DF, Robinson MS, Noebels JL, Burmeister ML. Genetic and phenotypic analysis of the mouse mutant mh2J, an Ap3d allele caused by IAP element insertion. Mamm Genome 2003; 14: 157-67. PMID: 12647238
- Kantheti P, Qiao X, Diaz ME, Peden AA, Meyer GE, Carskadon SL, Kapfhamer D, Sufalko D, Robinson MS, Noebels JL, Burmeister M. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron 1998; 21: 111-22. PMID: 9697856
- Odorizzi G, Cowles CR, Emr SD. The AP-3 complex: a coat of many colours. Trends Cell Biol 1998; 8: 282-8. PMID: 9714600
- Ooi CE, Dell'Angelica EC, Bonifacino JS. ADP-Ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J Cell Biol 1998; 142: 391-402. PMID: 9679139
- Ooi CE, Moreira JE, Dell'Angelica EC, Poy G, Wassarman DA, Bonifacino JS. Altered expression of a novel adaptin leads to defective pigment granule biogenesis in the Drosophila eye color mutant garnet. EMBO J 1997; 16: 4508-18. PMID: 9303295
- Sugita M, Cao X, Watts GF, Rogers RA, Bonifacino JS, Brenner MB. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity 2002; 16: 697-706. PMID: 12049721
EDIT HISTORY:
Created by Wei Li: 07/02/2004